AWS HealthOmics에서 Annotation 작업 수행하기
AWS HealthOmics의 Analytics 기능을 활용하여 annotation작업을 수행할 수 있습니다.
준비물
- 입력 샘플 VCF
- Annotation할 정보 소스 VCF (예: ClinVar)
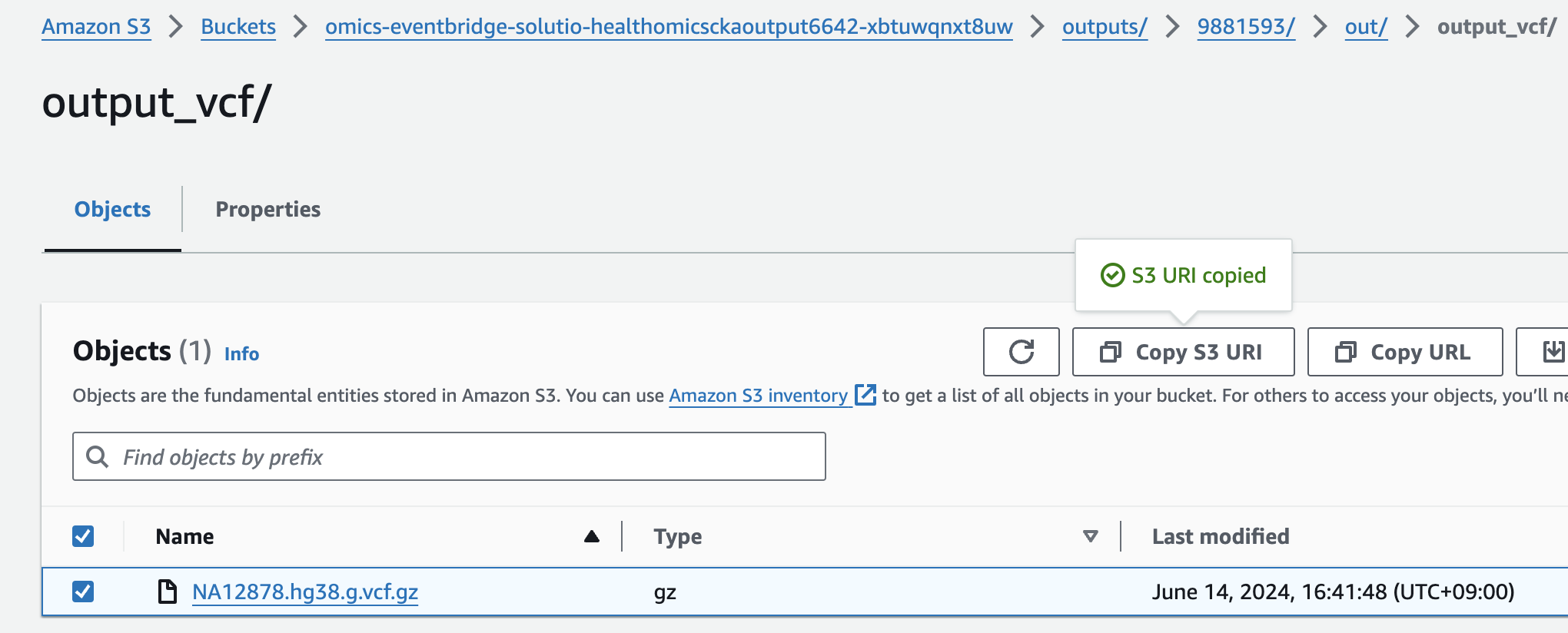
s3://omics-eventbridge-solutio-healthomicsckaoutput6642-xbtuwqnxt8uw/outputs/9881593/out/output_vcf/NA12878.hg38.g.vcf.gz
Variant stores
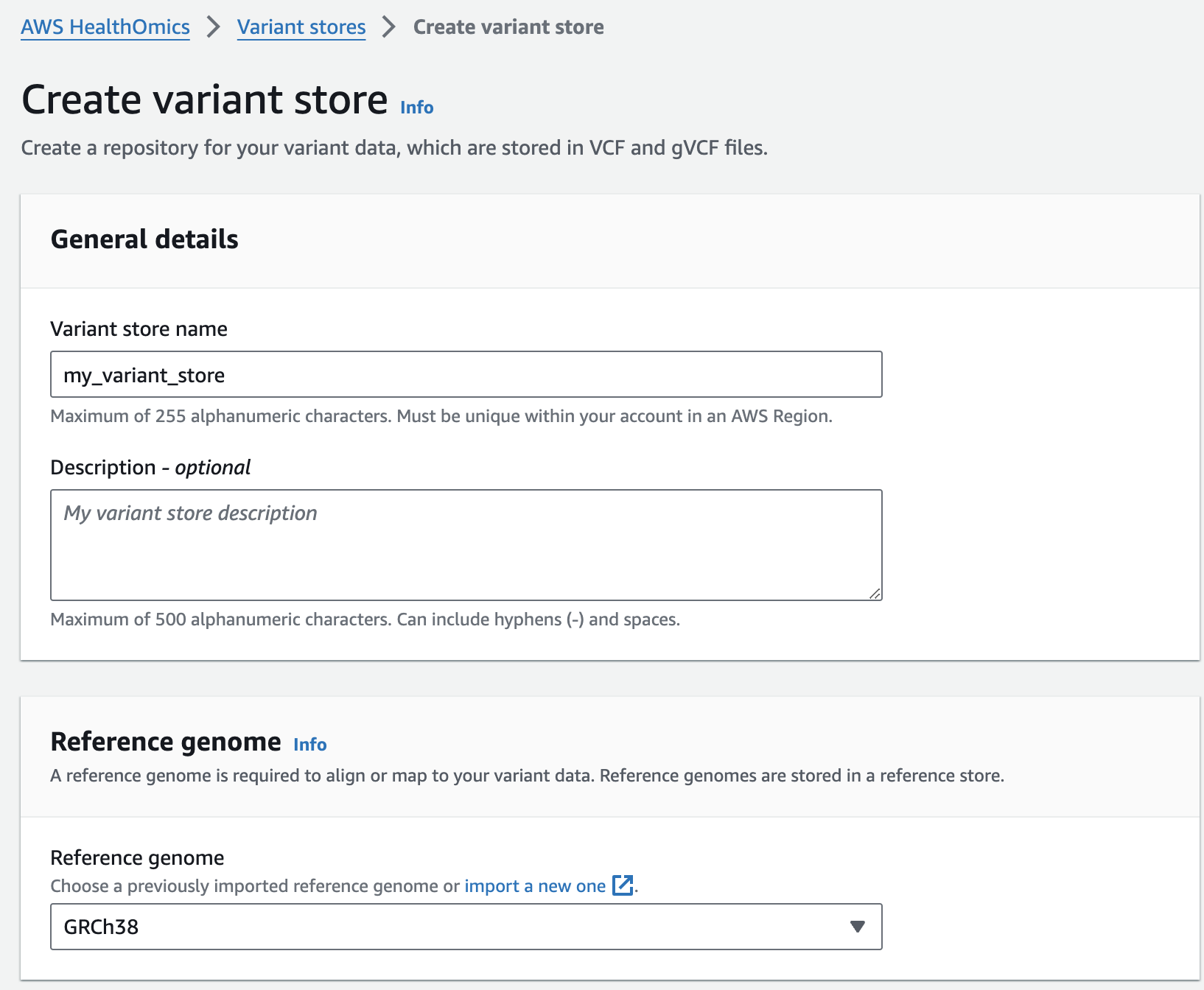
변이 스토어 생성
- From the AWS HealthOmics Console, navigate to Analytics > Variant stores
- Select Create variant store
- For Variant store name provide "my_variant_store".
- For Reference genome select "GRCh38" (this is a pre-provisioned reference, but you can alternatively select the reference you imported in the Reference Store part of the workshop)
- Finish with Create variant store
변이스토어에 샘플 VCF 파일 가져오기
Next, you are going to start a VCF import job. To do this:
- From the AWS HealthOmics Console, navigate to Analytics > Variant stores
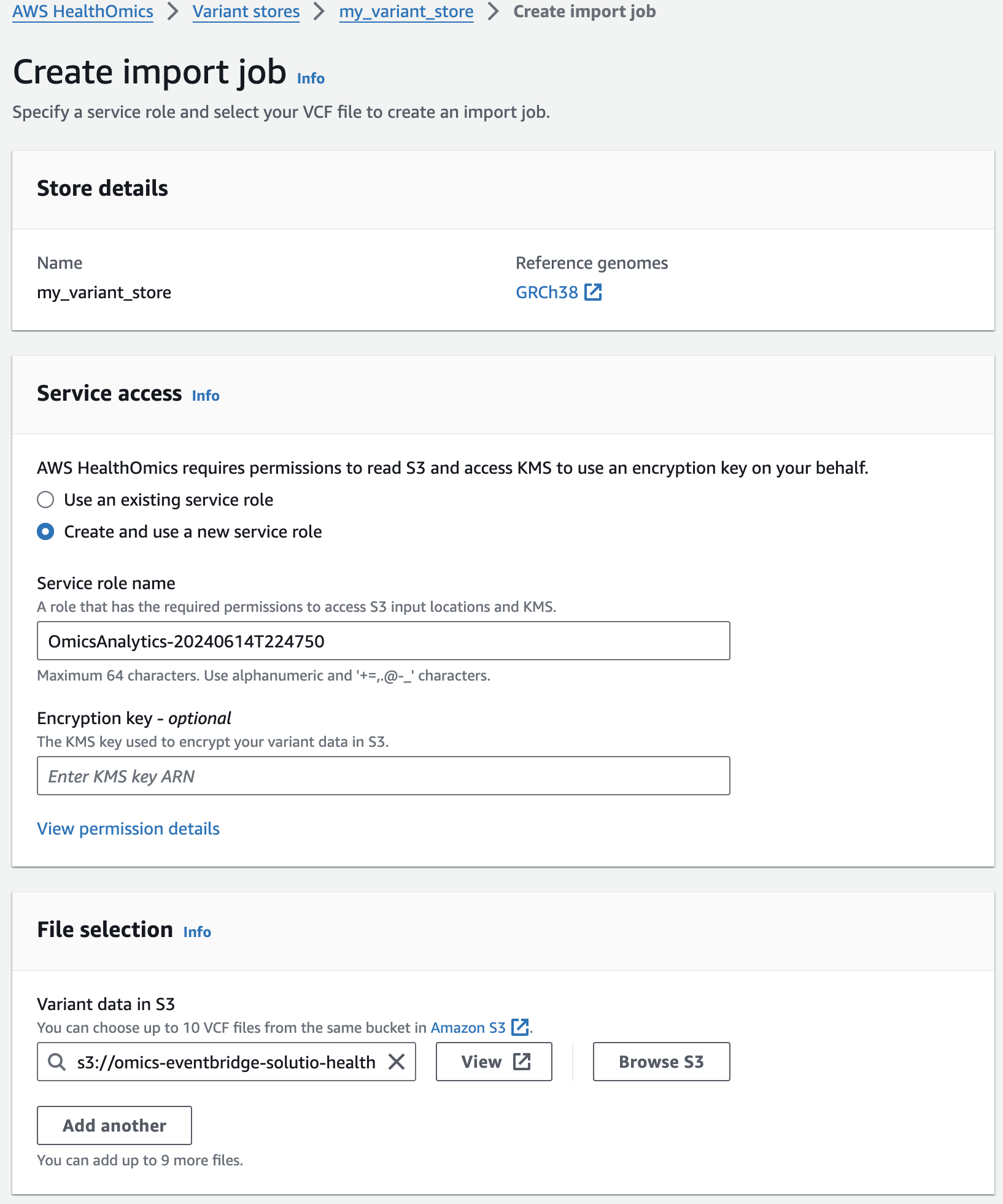
- Select the Name Variant store named omicsvariantstore1 (or the one you created above as appropriate)
- Select Import variant data. If this option isn't available select Actions > Import.
- Select Create and use a new service role
- For Select variant data from S3 provide the following S3 URI:
아래 s3 경로는 입력 VCF 파일의 S3 URI을 의미합니다.
s3://omics-eventbridge-solutio-healthomicsckaoutput6642-xbtuwqnxt8uw/outputs/9881593/out/output_vcf/NA12878.hg38.g.vcf.gzNOTE: The region will differ based on deployment region.
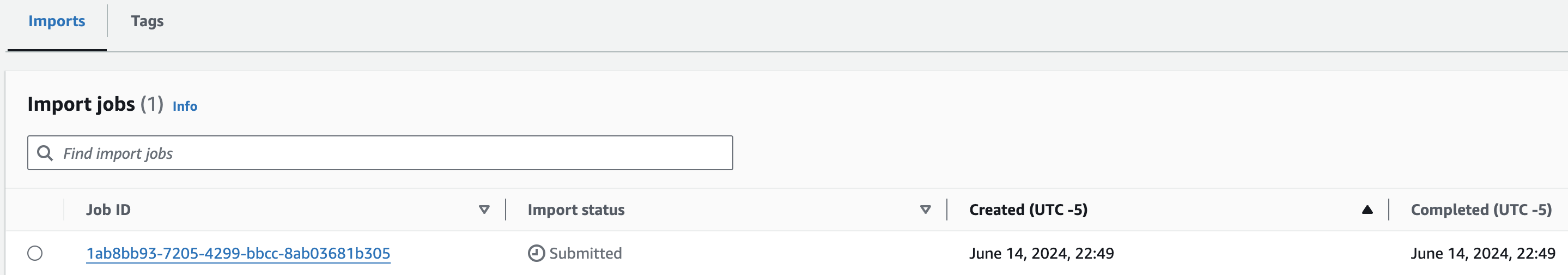
- Start the import with Create import job
You should now see something like this:
- 적당한 Service role 이 없을 경우 새로 생성하여 사용하는 옵션을 선택할 수 있습니다.
- 앞에서 설명한대로 입력하고자하는 VCF 파일의 S3 경로를 작성합니다.
콘솔에서 VCF Import작업시 제출되었음을 확인할 수 있습니다.
Annotation stores
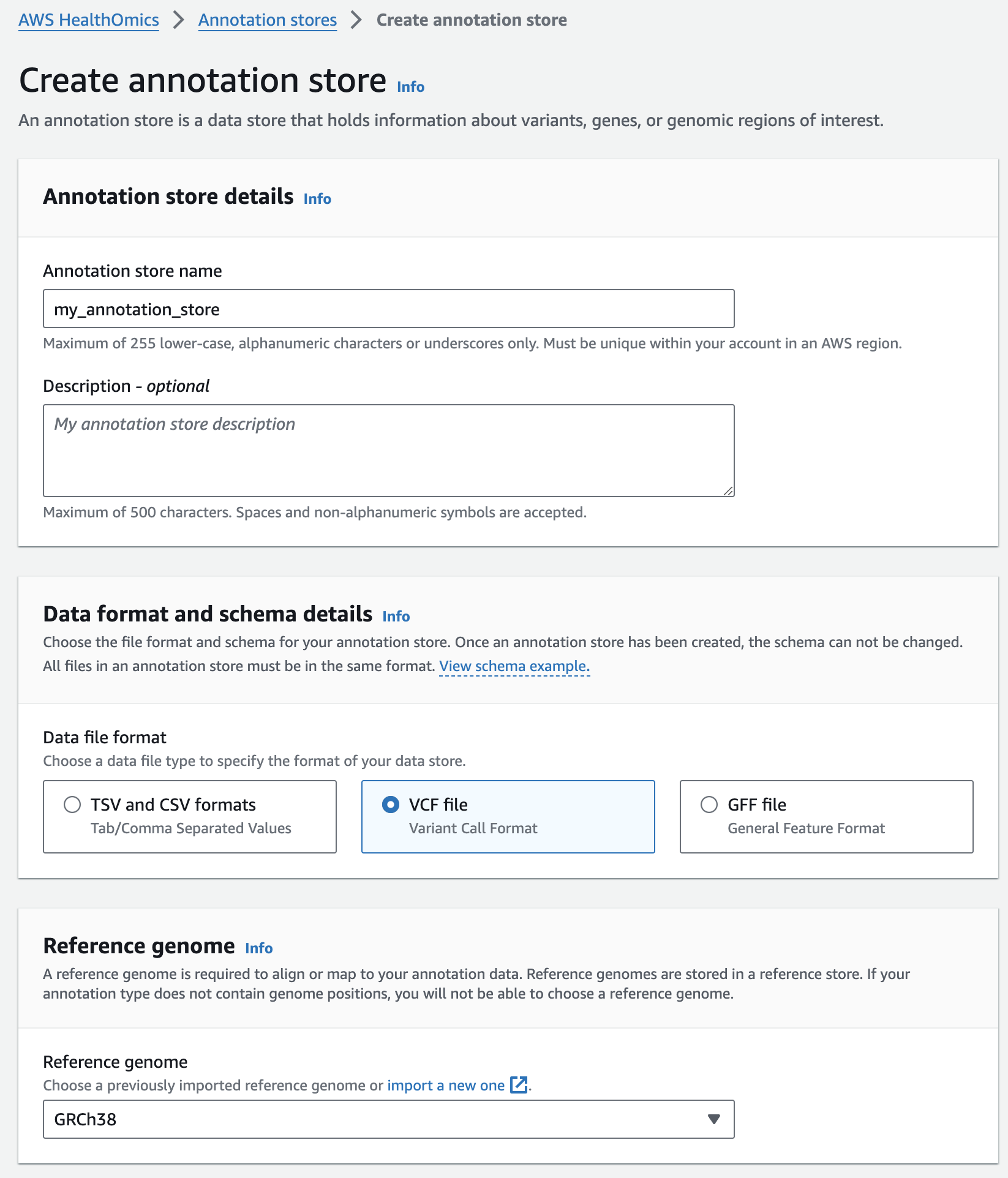
주석 스토어 생성
- From the AWS HealthOmics Console, navigate to Analytics > Annotation stores
- Select Create annotation store
- For Variant store name provide "my_annotation_store".
- For Data file format select VCF file
- For Reference genome select "GRCh38" (this is a pre-provisioned reference, but you can alternatively select the reference you imported in the Reference Store part of the workshop)
- Finish with Create annotation store
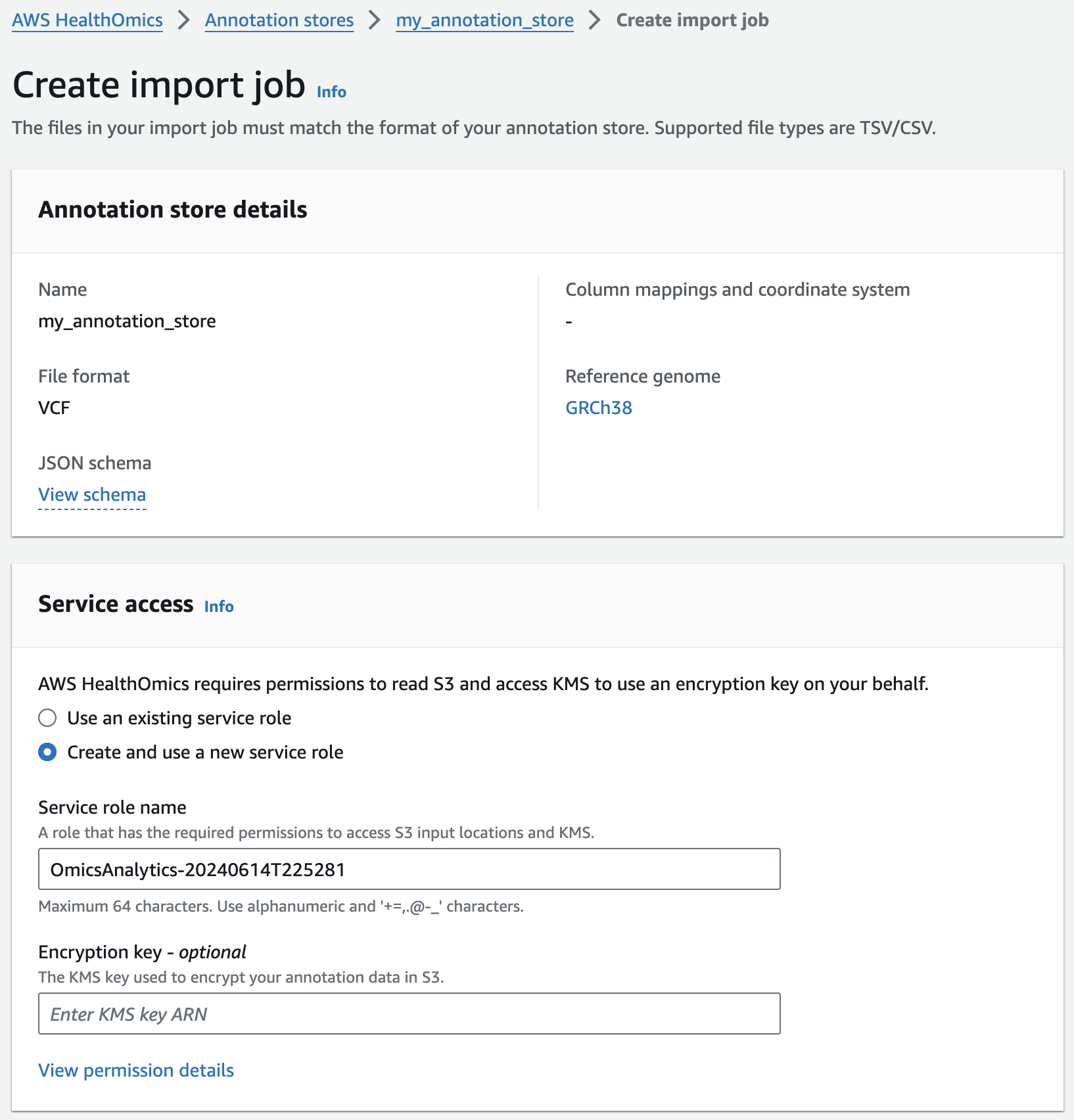
주석 스토어에 VCF 파일 가져오기
Next, you are going to start an annotation import job to import ClinVar annotations in VCF format into the pre-provisioned store.
- From the AWS HealthOmics Console, navigate to Analytics > Annotation stores
- Click on the name of the Annotation store named omicsannotationstore1 (or the one you created above as appropriate)
- Under Store versions click on the name of the only version listed (there should only be one at this time).
- Select Import VCF data. If this option isn't available select Actions > Import.
- Select Create and use a new service role
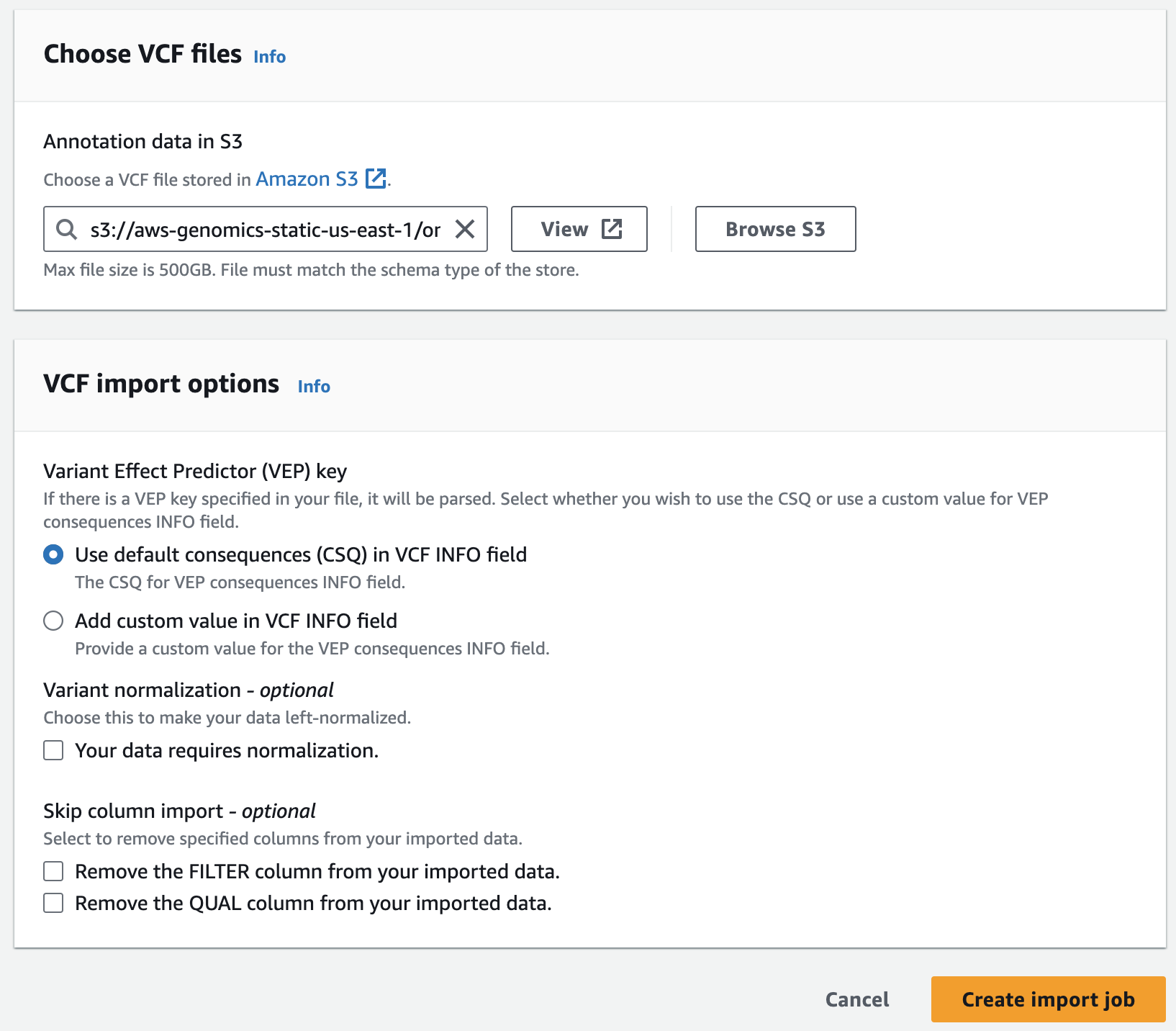
- For Choose annotation data from S3 provide the following S3 URI, including the appropriate AWS region:
아래는 미리준비된 예제 clinvar 입니다.
s3://aws-genomics-static-<aws-region>/omics-workshop/data/annotations/clinvar.vcf.gz실제 clinvar 데이터는 여기서 다운로드 할 수 있습니다.
예: https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh38/
Querying variants and annotations
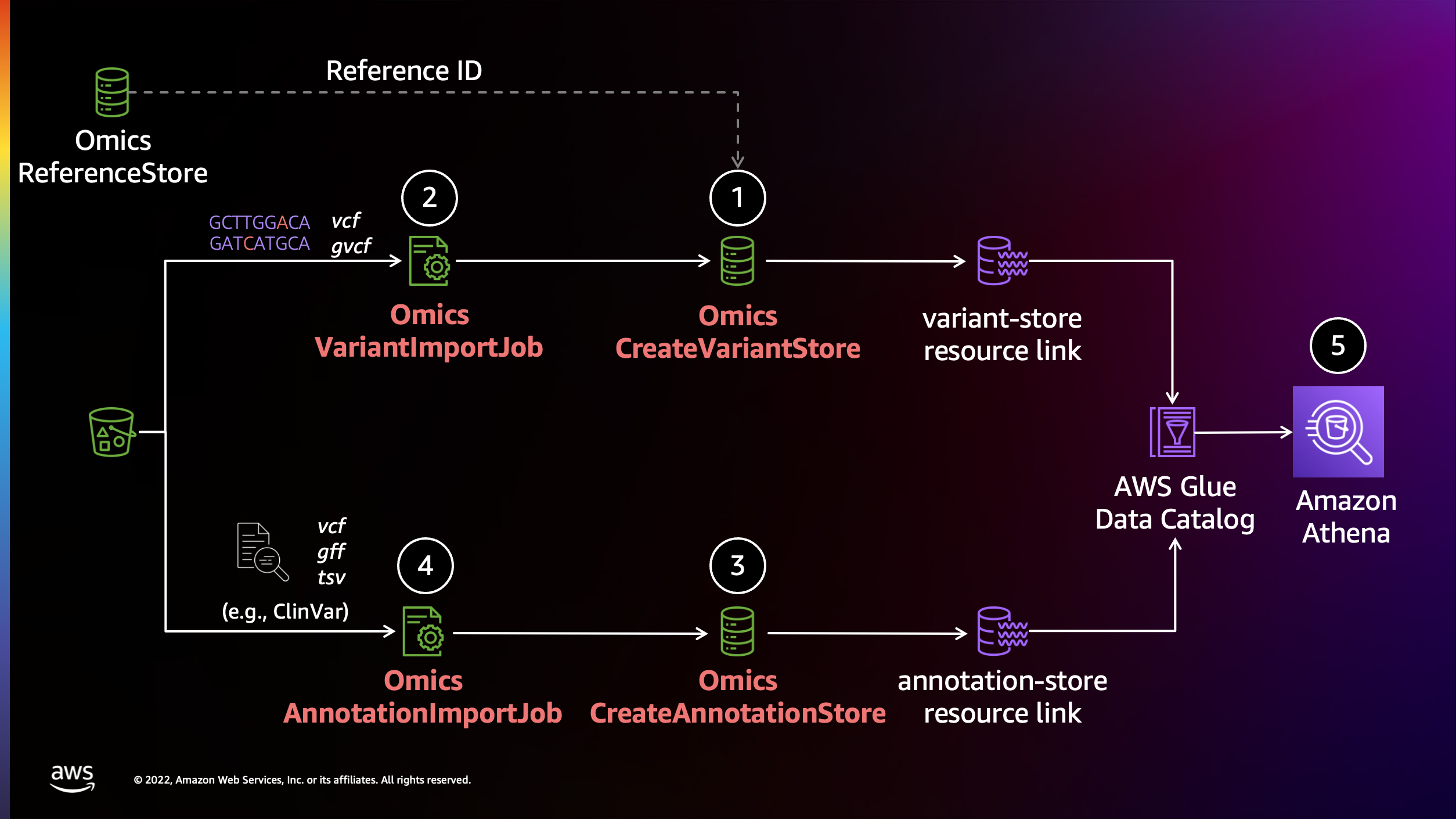
이전 섹션에서 가져온 variant 및 annotation 데이터는 확장 가능한 쿼리가 가능한 열 형식의 저장소(Apache Parquet)로 변환됩니다. 데이터는 AWS 레이크 형성에서 공유 데이터베이스 및 테이블로 사용할 수 있습니다. 데이터를 쿼리하기 전에 Lake Formation 리소스 링크를 통해 액세스 권한을 제공하고 Athena 작업 그룹을 만드는 등 몇 가지 설정 단계를 수행해야 합니다.
- AWS Lake Formation을 사용하여 데이터 레이크 관리자 및 리소스 링크 생성하기
- Amazon Athena에서 작업 그룹을 생성하고 쿼리 편집기를 사용하여 변형 및 어노테이션 저장소에 대한 간단한 쿼리 실행하기
- AWS SDK for Pandas (aka AWS Wrangler)를 사용해 SageMaker 노트북에서 변형 및 어노테이션 저장소에 대해 쿼리 실행하기
AWS Lake Formation 서비스 셋업
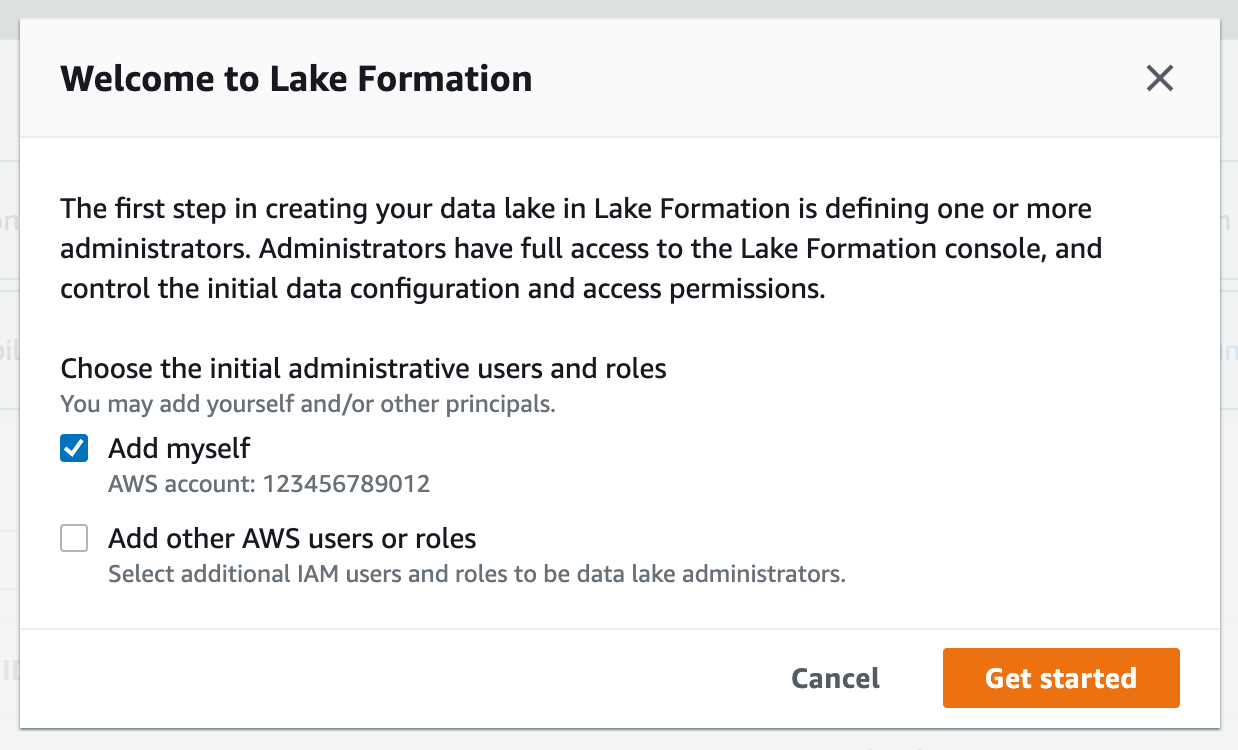
Lake Formation Data Lake administrators are users and roles with permissions to create resource links (covered in the section below). In a real-world scenario, you would only need to setup data lake administrators once per account per region, or you would have IT support staff that serve this role.
For this workshop, you will need to verify that your current user role is a data lake administrator.
2. If you see the following screen, select Get started:

4. If WSParticipantRole is not listed as a data lake administrator, select Choose administrators and then choose WSParticipantRole under IAM uesrs and roles. Then select Save to add the role as a Data lake administrator.
데이터베이스 생성
Let's create a database that we'll use as a virtual container for our variants and annotations.
1. AWS Lake Formation 콘솔에서 Databases 로 들어갑니다.
2. Select Create Database.
3. For Name provide omicsdb. 
4. Accept all other defaults and finish with Create database.
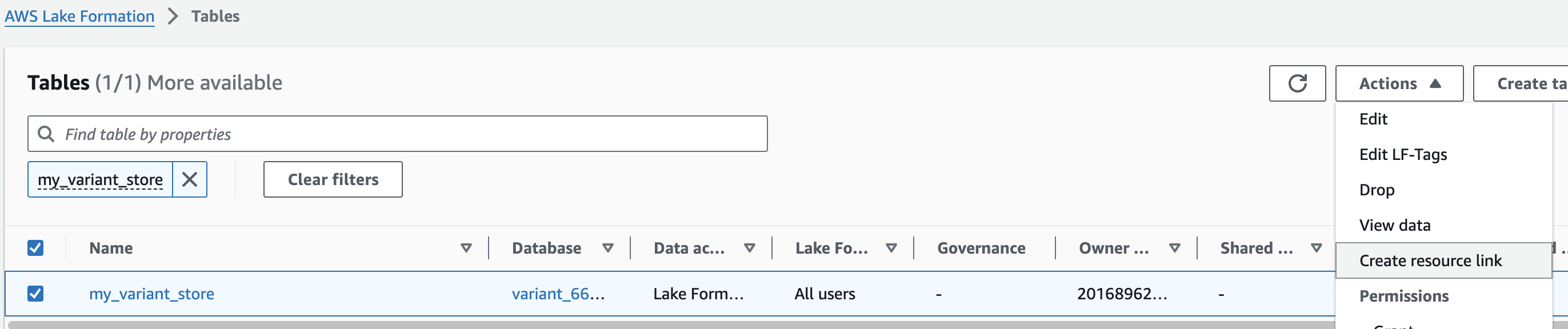
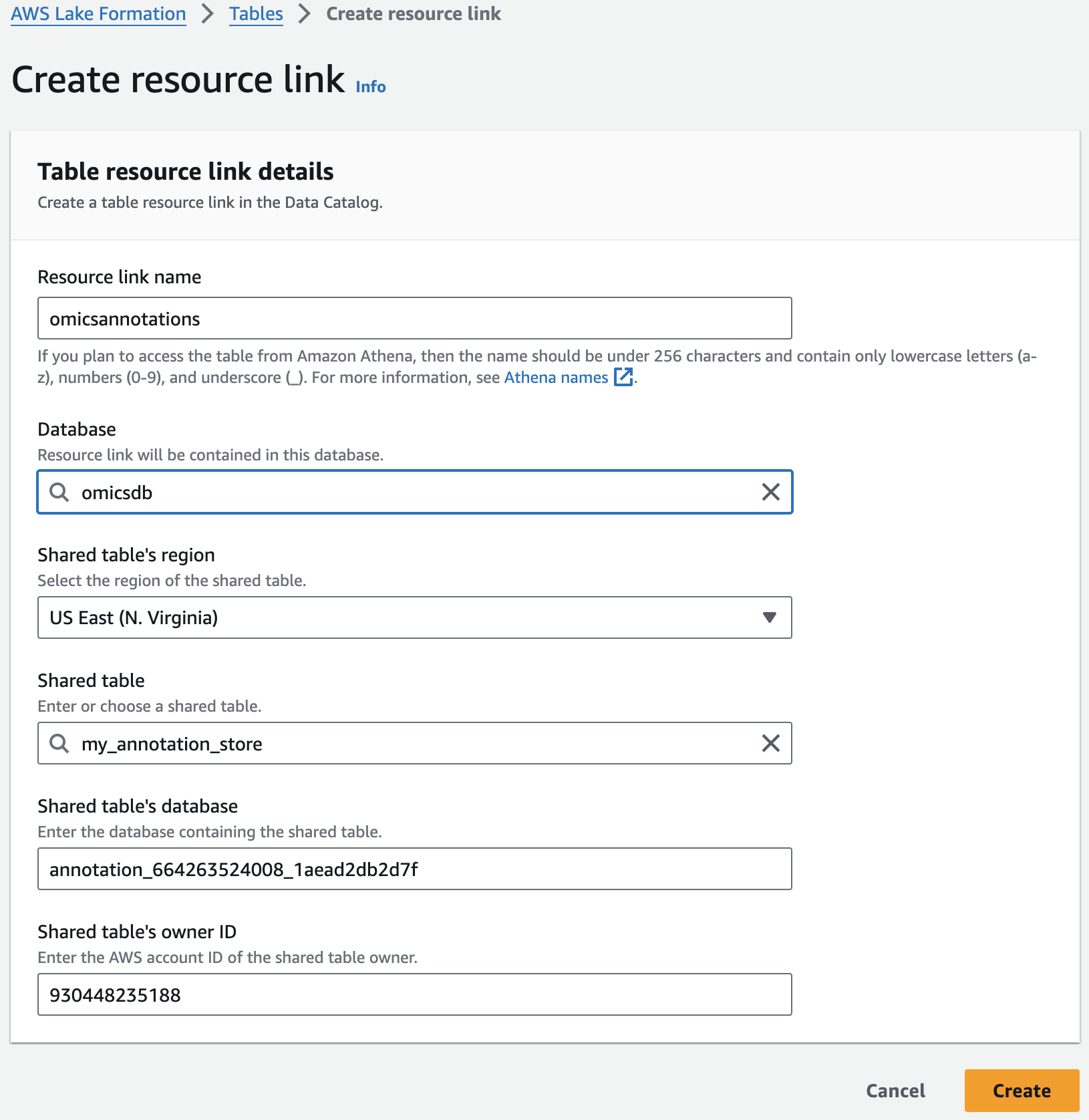
리소스 링크 생성
Resource links connect resources shared by HealthOmics Analytics to new or existing databases in your AWS Glue Data Catalog. For this workshop, we'll create resource links within the omicsdb you created above that point to the Variant and Annotaiton stores you created in previous sections.
1. AWS Lake Formation 콘솔에서 Tables 메뉴로 진입합니다.
2. 앞에서 만들었던 변이 스토어 이름을 검색한 뒤 선택하고 새로운 리소스 링크를 만듭니다.
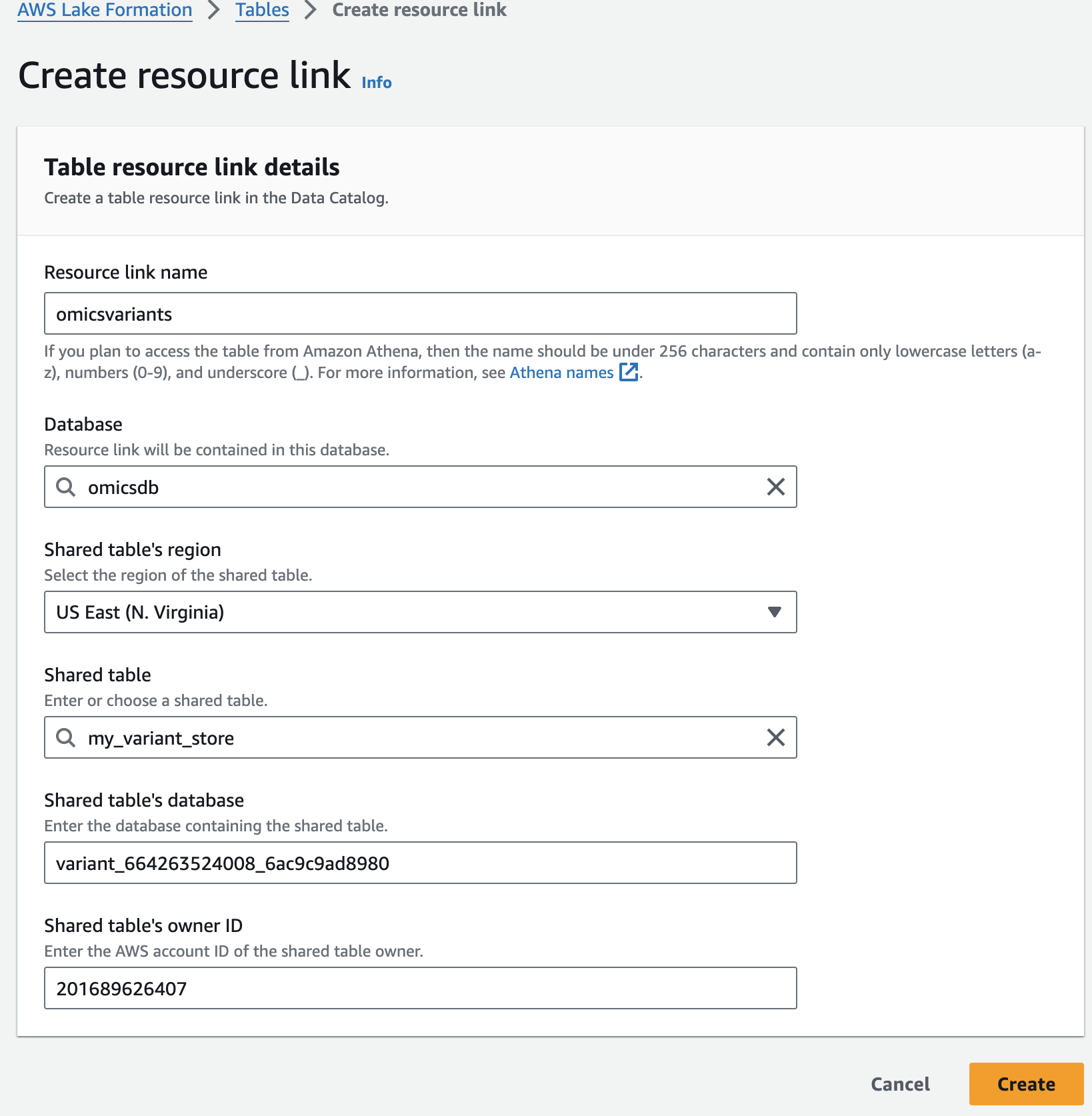
여기 예는 my_variant_store 입니다.
For Resource link name, provide omicsvariants.
For Database, provide omicsdb.
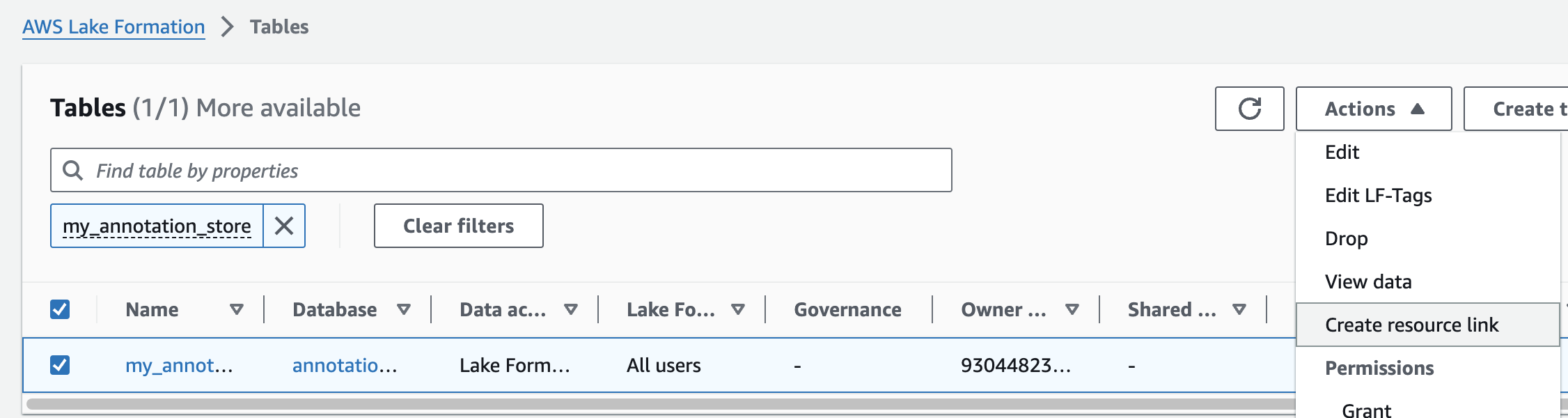
주석 테이블에 대해 위의 단계를 반복하여 omicsdb 데이터베이스에 omicsannotations라는 리소스 링크를 만듭니다.
Amazon Athena 셋업
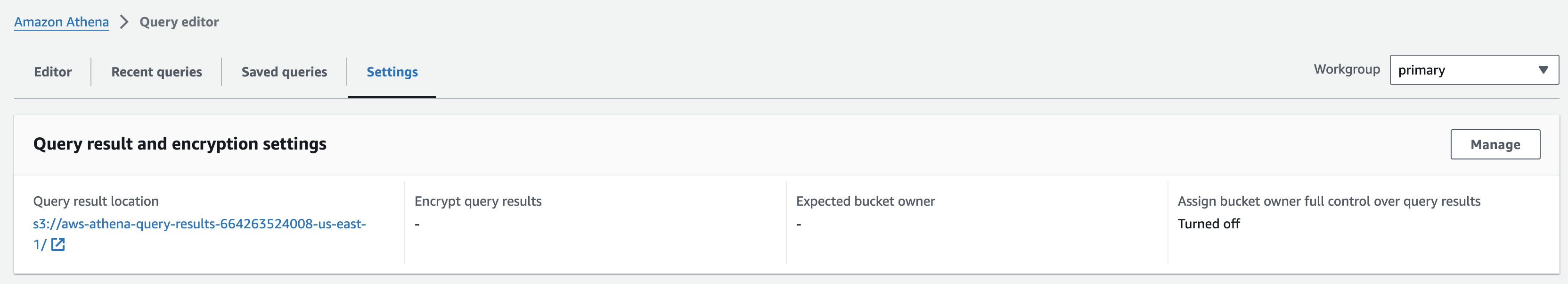
쿼리 결과 디렉토리 지정
- Open the Amazon Athena console .
- From the navigation, choose Query editor.
- In the Query editor, choose the Settings tab and then choose Manage.
- Click on Browse S3, select the bucket named omics-output-{REGION}-{ACCOUNT-ID}, and click Choose. This will fill the Location of query result with the S3 URI of the workshop output bucket. Append to this URI "/athena/" and select Save.
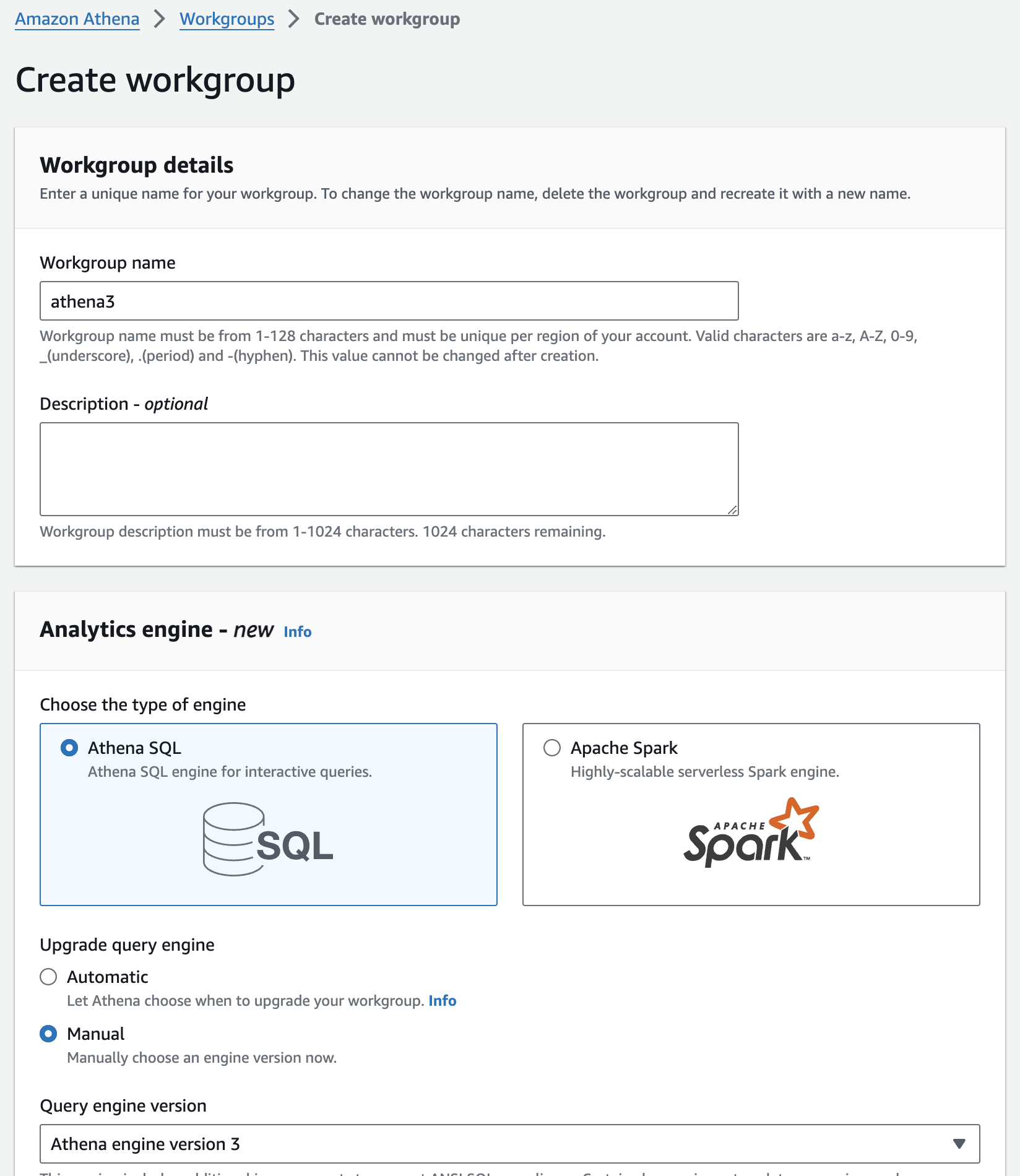
워킹 그룹 생성
- Open the Amazon Athena console .
- From the navigation, choose Workgroups, and then Create Workgroup.
- For Workgroup name provide athena3.
- Select Athena SQL for the type of engine.
- Under Upgrade query engine select Manual.
- Under Query Engine Version select Athena version 3.
- Finish with Create workgroup
Running queries - Amazon Athena
Now that we have Athena configured, let's run some queries.
- Open the Amazon Athena Query editor .
- Under Workgroup select "athena3" (which you created above).
- Make sure the Data Source is AwsDataCatalog and the Database is omicsdb (which you created previously). Both the omicsvariants and omicsannotations tables should be listed.
간단한 쿼리
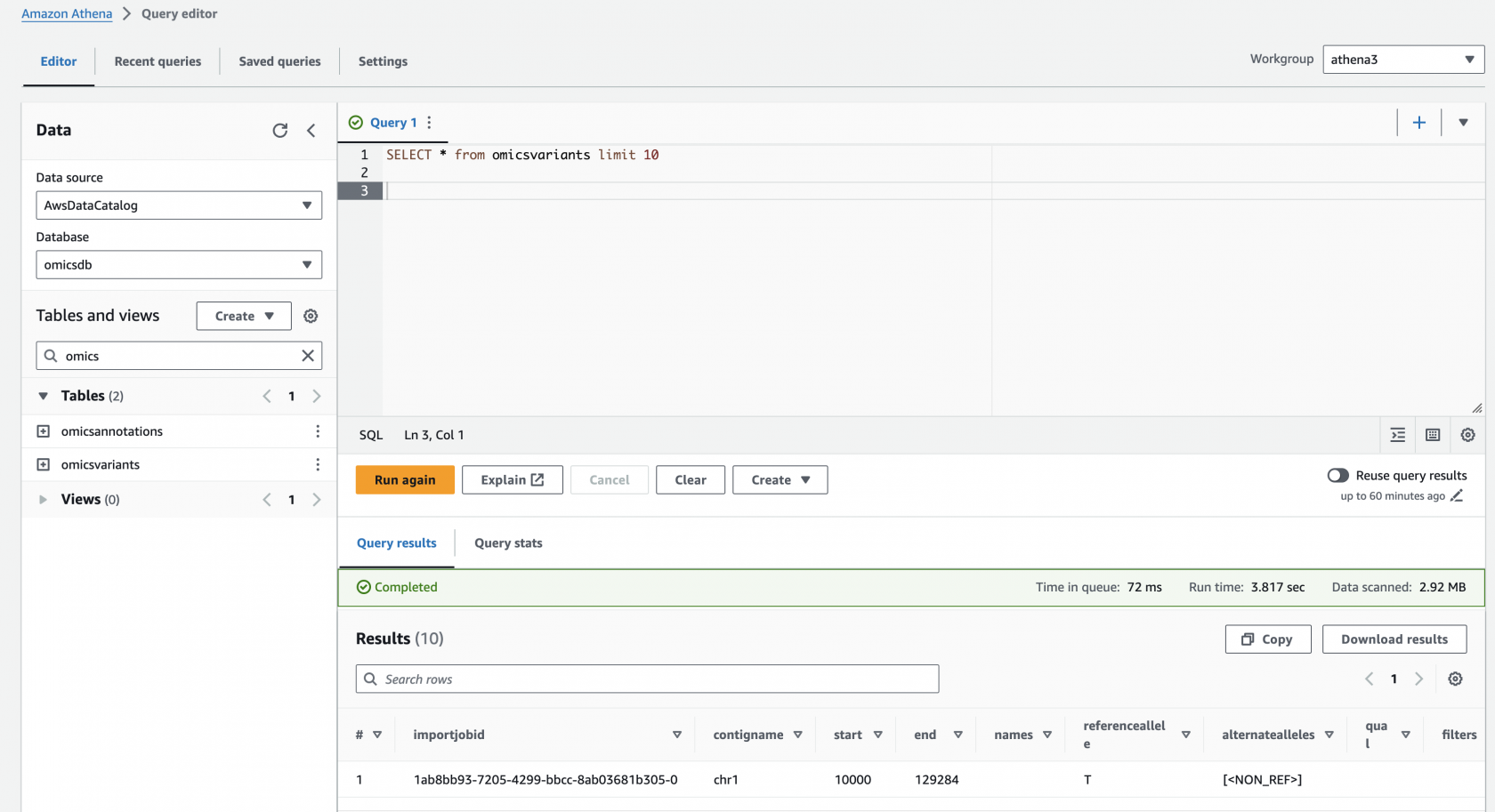
Preview the omicsvariants table, by running the following query:
SELECT * from omicsvariants limit 10- Copy the above query and paste it into the Query Editor under the Query 1 tab.
- Select Run to execute the query.
Results should return in a few seconds and look like:
복잡한 쿼리
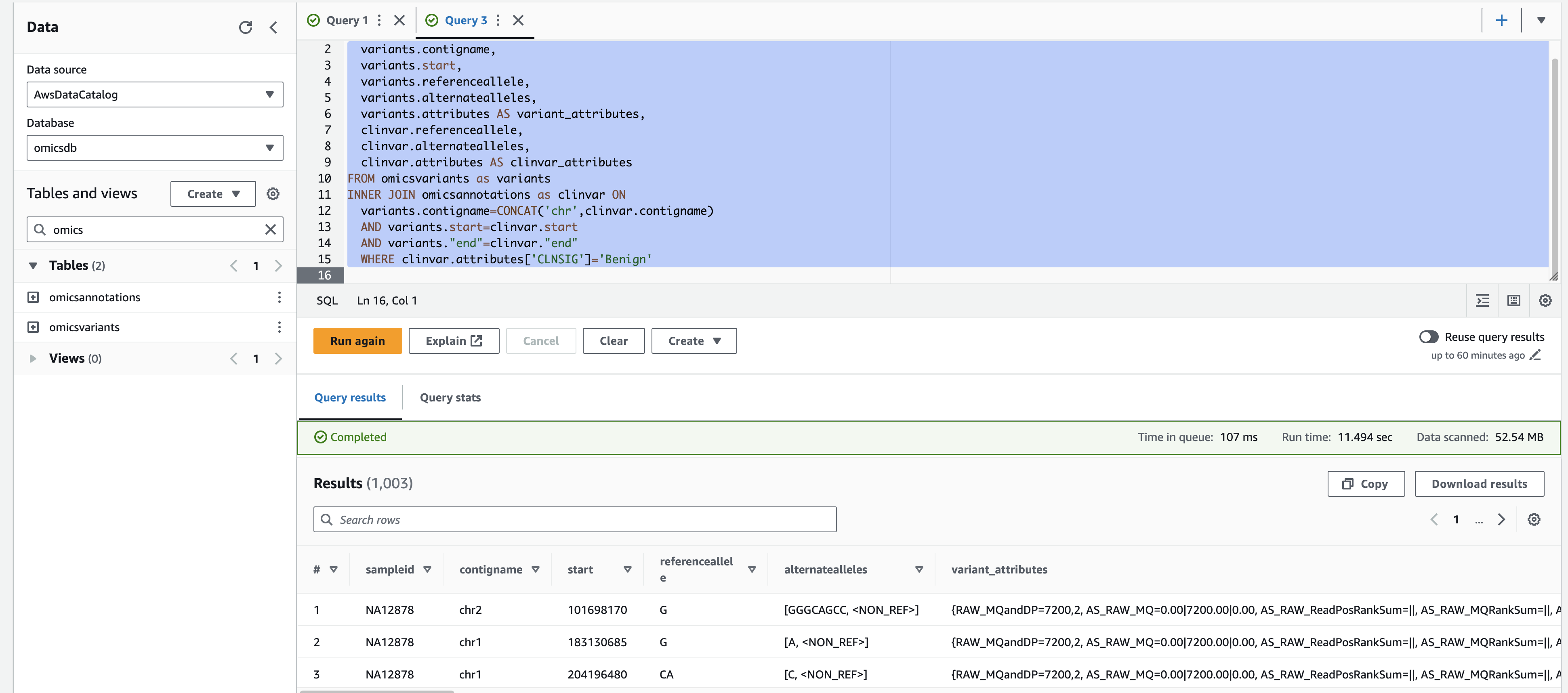
For a more complex query, you can run the following which displays variants with a 'Likely_pathogenic' clinical significance by joining ClinVar annotations to variants.
- Select the + on the top right to create a new query tab called Query 2.
- Copy and paste the SQL below and select Run.
SELECT variants.sampleid,
variants.contigname,
variants.start,
variants.referenceallele,
variants.alternatealleles,
variants.attributes AS variant_attributes,
clinvar.referenceallele,
clinvar.alternatealleles,
clinvar.attributes AS clinvar_attributes
FROM omicsvariants as variants
INNER JOIN omicsannotations as clinvar ON
variants.contigname=CONCAT('chr',clinvar.contigname)
AND variants.start=clinvar.start
AND variants."end"=clinvar."end"
WHERE clinvar.attributes['CLNSIG']='Benign'
또다른 예 (본인의 상황에 맞게 수정해야할 것입니다.)
SELECT variants.sampleid,
variants.contigname,
variants.start,
variants.referenceallele,
variants.alternatealleles,
variants.attributes AS variant_attributes,
clinvar.attributes AS clinvar_attributes
FROM omicsvariants as variants
INNER JOIN omicsannotations as clinvar ON
variants.contigname=CONCAT('chr',clinvar.contigname)
AND variants.start=clinvar.start
AND variants."end"=clinvar."end"
AND variants.referenceallele=clinvar.referenceallele
AND variants.alternatealleles=clinvar.alternatealleles
WHERE clinvar.attributes['CLNSIG']='Likely_pathogenic'
참고
- https://catalog.workshops.aws/amazon-omics-end-to-end/en-US/010-xp-console/300-omics-analytics
- https://github.com/aws-samples/amazon-omics-tutorials/blob/main/notebooks/200-omics_analytics.ipynb
- https://github.com/vcflib/vcflib/tree/master
- https://github.com/Ensembl/ensembl-vep
- echtvar
- https://github.com/brentp/vcfanno

No comments to display
No comments to display