AWS HealthOmics로 Nf-core 워크플로우 마이그레이션 하기 (scrnaseq)
참고문서: https://catalog.us-east-1.prod.workshops.aws/workshops/76d4a4ff-fe6f-436a-a1c2-f7ce44bc5d17/en-US
이 문서를 참고하여 환경을 준비합니다. 여기서는 이 과정은 생략합니다.
2025년 4월 18일 기준 scrnaseq 버전 History 2.7.1 > 3.0.0 > 4.0.0
현재 아래 내용은 2.7.0, 2.7.1에서만 테스트되었습니다.
프로젝트 셋업
버킷 생성 및 Bash 환경변수 선언
cd ~
export yourbucket="your-bucket-name"
export your_account_id="your-account-id"
export region="your-region"
export workflow_name="your-workflow-name"
export omics_role_name="your_omics_rolename"
# if not exist the bucket, let's create.
#aws s3 mb $yourbucket
nf-core repository로부터 워크플로우 복제
cd ~
git clone https://github.com/nf-core/scrnaseq --branch 2.7.1 --single-branchDocker Image Manifest의 생성
cp ~/amazon-ecr-helper-for-aws-healthomics/lib/lambda/parse-image-uri/public_registry_properties.json namespace.configinspect_nf.py 를 실행합니다.
python3 amazon-omics-tutorials/utils/scripts/inspect_nf.py \
--output-manifest-file scrnaseq_271_docker_images_manifest.json \
-n namespace.config \
--output-config-file omics.config \
--region $region \
~/scrnaseq/
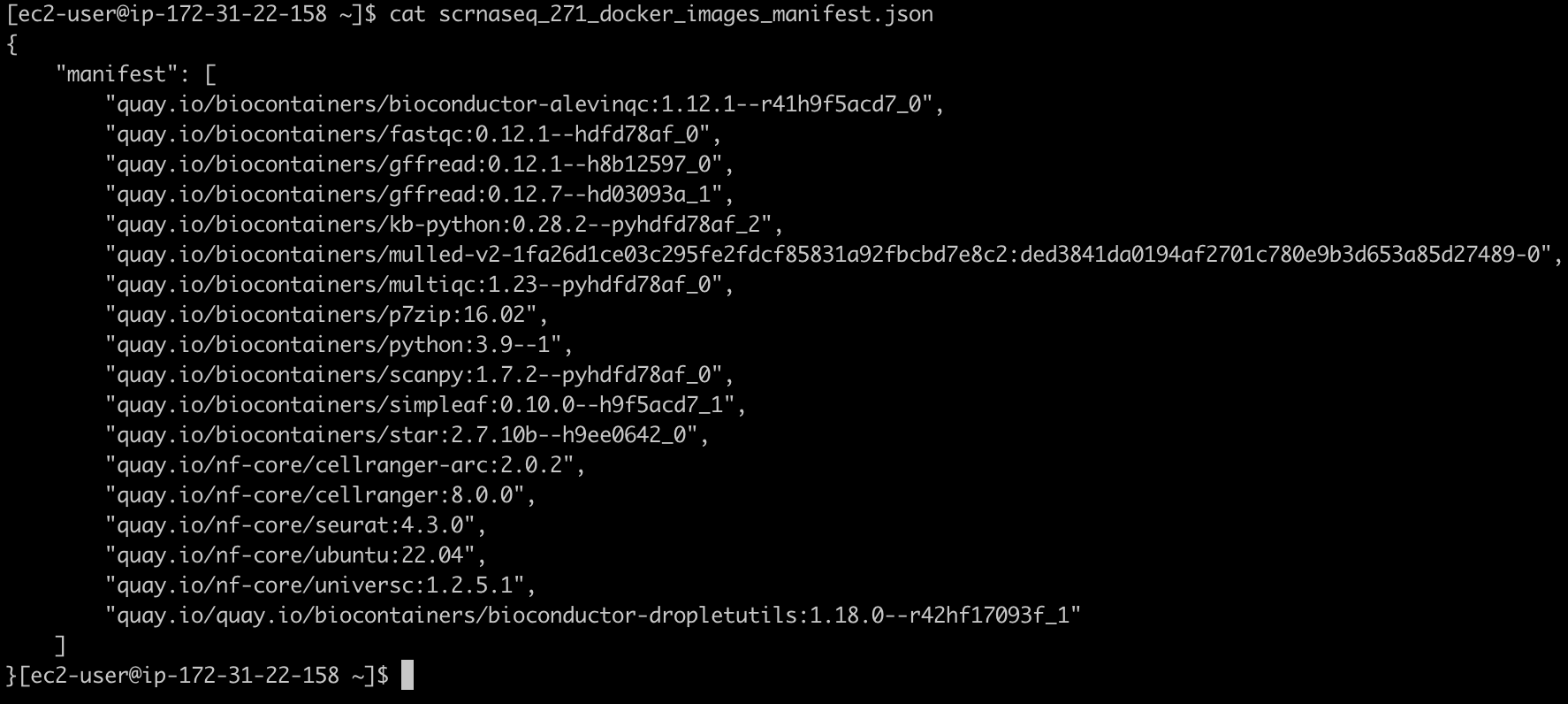
생성되는 두 개의 출력은 scrnaseq_271_docker_images_manifest.json과 omics.config입니다.
scrnaseq_271_docker_images_manifest.json 파일은 예를들어 다음과 같은 모습이어야 합니다:
컨테이너 사설화 (into Amazon ECR)
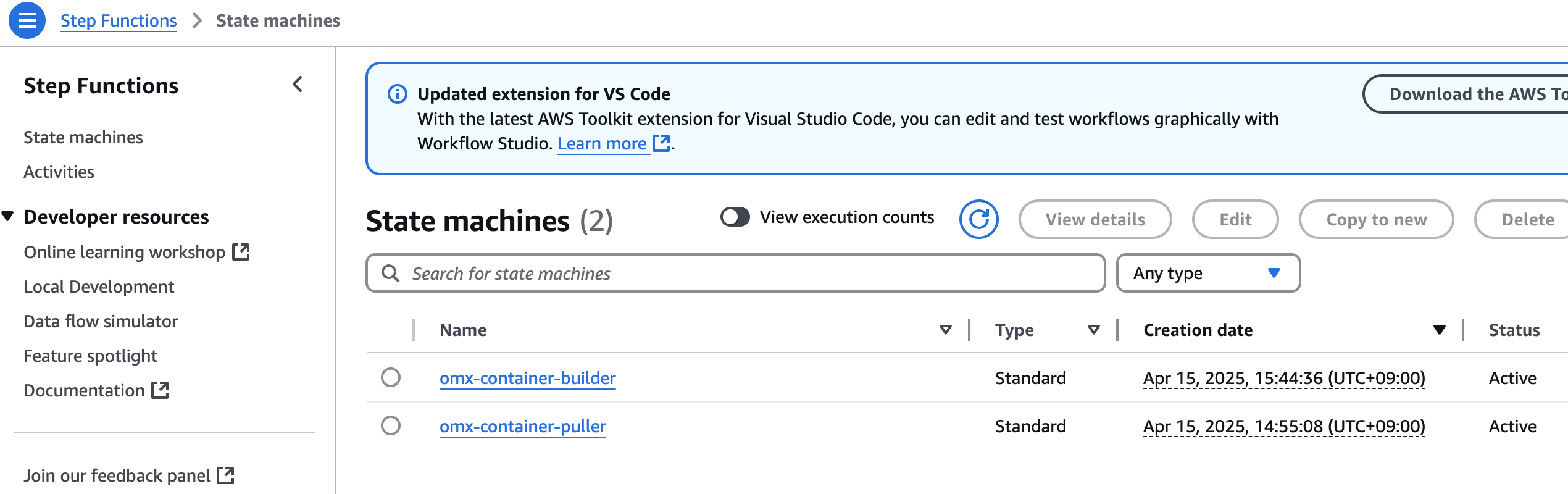
aws stepfunctions start-execution \
--state-machine-arn arn:aws:states:$region:$your_account_id:stateMachine:omx-container-puller \
--input file://scrnaseq_271_docker_images_manifest.json
step function 콘솔에서 state machines중에 omx-container-puller를 확인하여 Execution이 완료되었는지 확인합니다.
nf-core project 코드 업데이트
mv omics.config scrnaseq/conf
echo "includeConfig 'conf/omics.config'" >> scrnaseq/nextflow.config
AWS HealthOmics 워크플로우 만들기
단계1. AWS HealthOmics 파라미터 파일
parameter-description.json을 만들어 아래와 같이 저장합니다.
cat << EOF > parameter-description.json
{
"input": {"description": "Samplesheet with sample locations.",
"optional": false},
"protocol" : {"description": "10X Protocol used: 10XV1, 10XV2, 10XV3",
"optional": false},
"aligner": {"description": "choice of aligner: alevin, star, kallisto",
"optional": false},
"barcode_whitelist": {"description": "Optional whitelist if 10X protocol is not used.",
"optional": true},
"gtf": {"description": "S3 path to GTF file",
"optional": false},
"fasta": {"description": "S3 path to FASTA file",
"optional": false},
"skip_emptydrops": {"description": "module does not work on small dataset",
"optional": true}
}
EOF단계2. 워크플로우 스테이징
zip -r scrnaseq-workflow.zip scrnaseq -x "*/\.*" "*/\.*/**"
aws s3 cp scrnaseq-workflow.zip s3://${yourbucket}/workshop/scrnaseq-workflow.zip
aws omics create-workflow \
--name ${workflow_name} \
--definition-uri s3://${yourbucket}/workshop/scrnaseq-workflow.zip \
--parameter-template file://parameter-description.json \
--engine NEXTFLOW
단계3. 워크플로우 생성 확인
workflow_id=$(aws omics list-workflows --name ${workflow_name} --query 'items[0].id' --output text)
echo $workflow_id워크플로우 테스트하기
작은 크기의 예제 샘플
입력파일 준비
parameter-description.json에 사용된 것과 동일한 키를 사용하여 input.json 파일을 새로 만듭니다. 값은 워크플로에서 허용되는 실제 S3 경로 또는 문자열이 됩니다.
아래는 테스트 샘플의 파라미터 예시입니다. (참고)
데이터 준비 참고
예제 데이터는 다음과 같이 다운로드 해볼 수 있습니다.
wget https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv
wget https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/GRCm38.p6.genome.chr19.fa
wget https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/gencode.vM19.annotation.chr19.gtf이제 현재 디렉토리에 다운로드 된 파일들을 버킷에 업로드할 수 있습니다.
aws s3 sync . s3://omics-output-us-east-1-462922227709/workflow_migration_workshop/nfcore-scrnaseq-v4.0.0/sample sheet 만들기
cat << EOF > samplesheet-2-0.csv
sample,fastq_1,fastq_2,expected_cells,seq_center
Sample_X,s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/Sample_X_S1_L001_R1_001.fastq.gz,s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/Sample_X_S1_L001_R2_001.fastq.gz,5000,"Broad Institute"
Sample_Y,s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/Sample_Y_S1_L001_R1_001.fastq.gz,s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/Sample_Y_S1_L001_R2_001.fastq.gz,5000,"CRG Barcelona"
Sample_Y,s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/Sample_Y_S1_L002_R1_001.fastq.gz,s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/Sample_Y_S1_L002_R2_001.fastq.gz,5000,"CRG Barcelona"
EOF위에서 만든 samplesheet를 s3로 복사
aws s3 cp samplesheet-2-0.csv s3://${yourbucket}/nfcore-scrnaseq/samplesheet-2-0.csv입력 json 만들기 (위 sample sheet경로가 아래 내용중 input에 값으로 들어가게됨)
cat << EOF > input.json
{
"input": "s3://${yourbucket}/nfcore-scrnaseq/samplesheet-2-0.csv",
"protocol": "10XV2",
"aligner": "star",
"fasta": "s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/GRCm38.p6.genome.chr19.fa",
"gtf": "s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/gencode.vM19.annotation.chr19.gtf",
"skip_emptydrops": true
}
EOF실제 크기의 샘플
(출처: 참고)
데이터 내 버킷내에 준비
aws s3 sync s3://ngi-igenomes/test-data/scrnaseq/ s3://${yourbucket}/test-data/scrnaseq/ --exclude "*" --include "pbmc8k_S1_L00*"
sample sheet 만들기
cat << EOF > samplesheet_2.0_full.csv
sample,fastq_1,fastq_2,expected_cells
pbmc8k,s3://${yourbucket}/test-data/scrnaseq/pbmc8k_S1_L007_R1_001.fastq.gz,s3://${yourbucket}/test-data/scrnaseq/pbmc8k_S1_L007_R2_001.fastq.gz,10000
pbmc8k,s3://${yourbucket}/test-data/scrnaseq/pbmc8k_S1_L008_R1_001.fastq.gz,s3://${yourbucket}/test-data/scrnaseq/pbmc8k_S1_L008_R2_001.fastq.gz,10000
EOF위에서 만든 samplesheet를 s3로 복사
aws s3 cp samplesheet_2.0_full.csv s3://${yourbucket}/nfcore-scrnaseq/samplesheet_2.0_full.csv입력 json 만들기 (위 sample sheet경로가 아래 내용중 input에 값으로 들어가게됨)
cat << EOF > input_full.json
{
"input": "s3://${yourbucket}/nfcore-scrnaseq/samplesheet_2.0_full.csv",
"protocol": "10XV2",
"aligner": "star",
"fasta": "s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/GRCm38.p6.genome.chr19.fa",
"gtf": "s3://aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/gencode.vM19.annotation.chr19.gtf",
"skip_emptydrops": true
}
EOFPolicy 준비
Prepare IAM service role to run AWS HealthOmics workflow
your-bucket-name, your-account-id, your-region을 모두 본인 환경에 맞게 수정하여 사용하세요.
omics_workflow_policy.json 만들기
cat << EOF > omics_workflow_policy.json
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Action": [
"s3:GetObject"
],
"Resource": [
"arn:aws:s3:::${yourbucket}/*",
"arn:aws:s3:::aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/*"
]
},
{
"Effect": "Allow",
"Action": [
"s3:ListBucket"
],
"Resource": [
"arn:aws:s3:::${yourbucket}",
"arn:aws:s3:::aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0",
"arn:aws:s3:::aws-genomics-static-${region}/workflow_migration_workshop/nfcore-scrnaseq-v2.3.0/*"
]
},
{
"Effect": "Allow",
"Action": [
"s3:PutObject"
],
"Resource": [
"arn:aws:s3:::${yourbucket}/*"
]
},
{
"Effect": "Allow",
"Action": [
"logs:DescribeLogStreams",
"logs:CreateLogStream",
"logs:PutLogEvents"
],
"Resource": [
"arn:aws:logs:${region}:${your_account_id}:log-group:/aws/omics/WorkflowLog:log-stream:*"
]
},
{
"Effect": "Allow",
"Action": [
"logs:CreateLogGroup"
],
"Resource": [
"arn:aws:logs:${region}:${your_account_id}:log-group:/aws/omics/WorkflowLog:*"
]
},
{
"Effect": "Allow",
"Action": [
"ecr:BatchGetImage",
"ecr:GetDownloadUrlForLayer",
"ecr:BatchCheckLayerAvailability"
],
"Resource": [
"arn:aws:ecr:${region}:${your_account_id}:repository/*"
]
}
]
}
EOF
echo "omics_workflow_policy.json 파일이 생성되었습니다."trust_policy.json 만들기
cat << EOF > trust_policy.json
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Principal": {
"Service": "omics.amazonaws.com"
},
"Action": "sts:AssumeRole",
"Condition": {
"StringEquals": {
"aws:SourceAccount": "${your_account_id}"
},
"ArnLike": {
"aws:SourceArn": "arn:aws:omics:${region}:${your_account_id}:run/*"
}
}
}
]
}
EOF
echo "trust_policy.json 파일이 생성되었습니다."IAM Role 생성
aws iam create-role --role-name ${omics_role_name} --assume-role-policy-document file://trust_policy.json
Policy document 생성
aws iam put-role-policy --role-name ${omics_role_name} --policy-name OmicsWorkflowV1 --policy-document file://omics_workflow_policy.json
워크플로우 실행
작은 샘플의 예제는 input.json, 큰 샘플의 예제는 input_full.json
aws omics start-run \
--name scrnaseq_workshop_test_run_1 \
--role-arn arn:aws:iam::${your_account_id}:role/${omics_role_name}\
--workflow-id ${workflow_id} \
--parameters file://input.json \
--output-uri s3://${yourbucket}/output/
No Comments