Quickstart Hail (Eng)
CloudFormation stack preparation
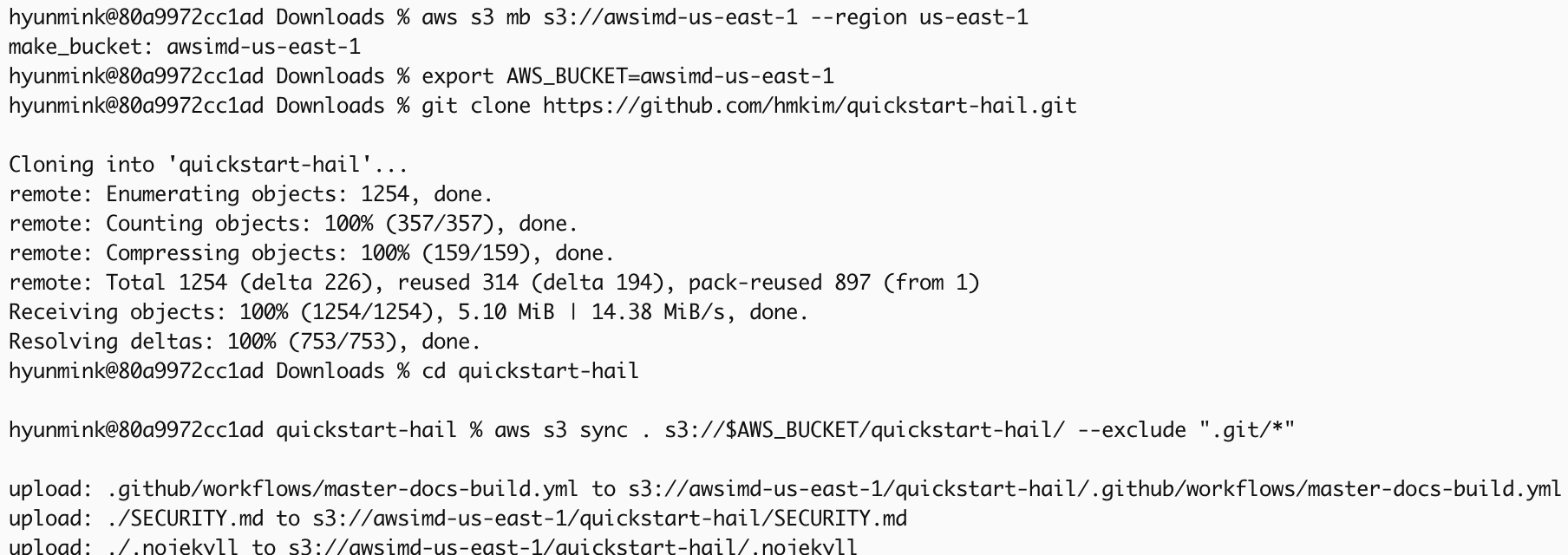
1. Prepare the AWS credentials and apply them in the terminal.
export AWS_DEFAULT_REGION="us-east-1"
export AWS_ACCESS_KEY_ID="{ACCESS_KEY}"
export AWS_SECRET_ACCESS_KEY="{SECRET_ACCESS_KEY}"
export AWS_SESSION_TOKEN="{SESSION_TOKEN}"2. Create an S3 bucket in the region where you want to launch this CloudFormation stack.
At this time, I recommend creating a bucket name using your own initial. As you know, the bucket already exists, it cannot be created.
aws s3 mb s3://{bucket name}-{region} --region {region}Download and unzip the content from this repository, then place the downloaded content into the S3 bucket you created earlier.
export AWS_BUCKET={bucket name}-{region}
git clone https://github.com/hmkim/quickstart-hail.git
cd quickstart-hail
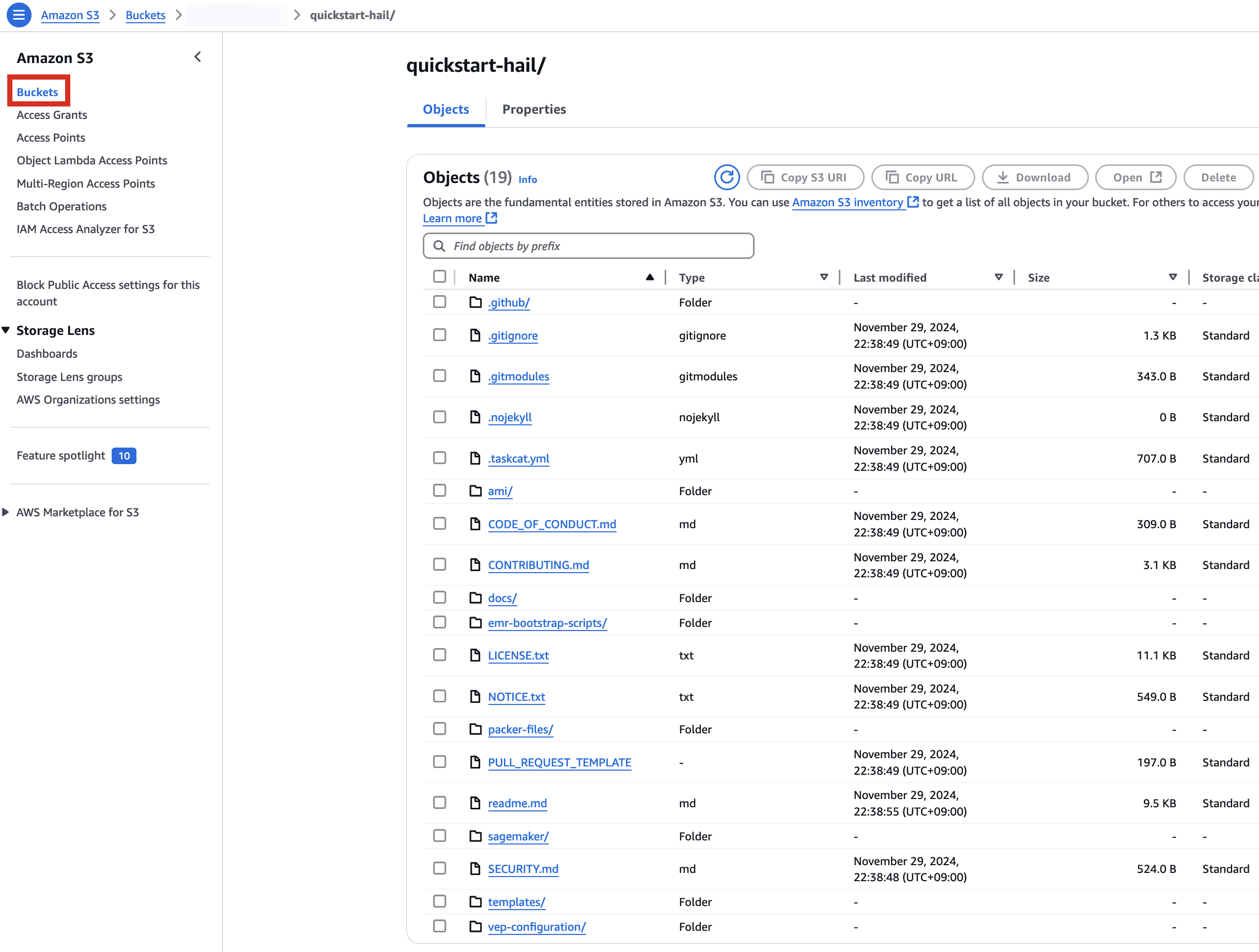
aws s3 sync . s3://$AWS_BUCKET/quickstart-hail/ --exclude ".git/*"3. Connect to the Amazon S3 console and check the bucket and directory.
Run the CloudFormation stack
1. Go to the CloudFormation console.
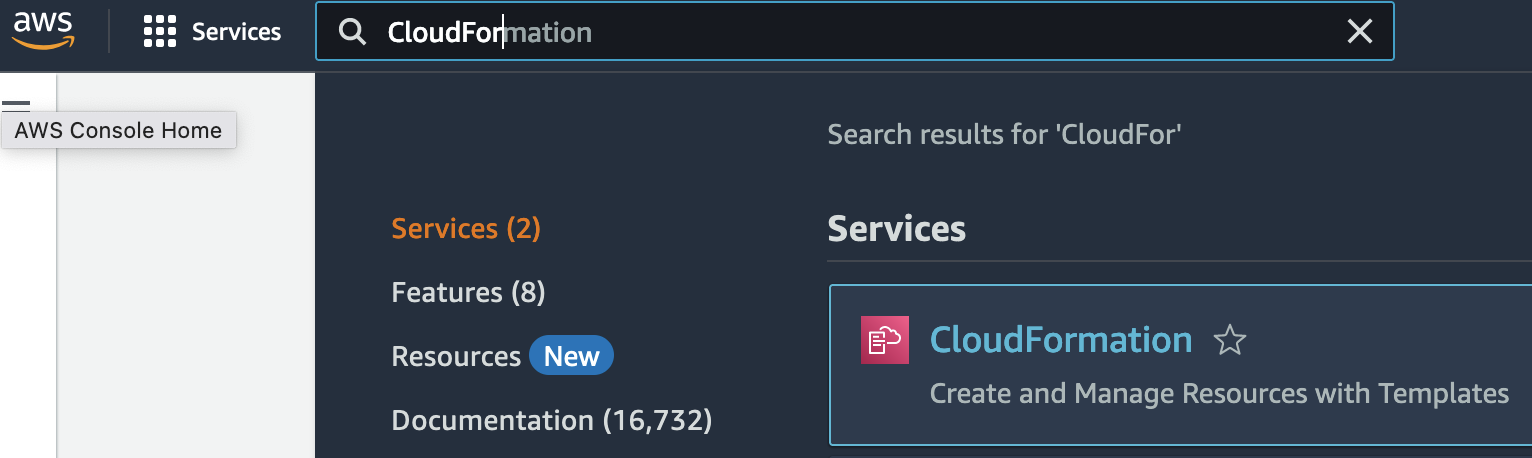
2. Creates a new stack. At this time, select With new resources (standard).
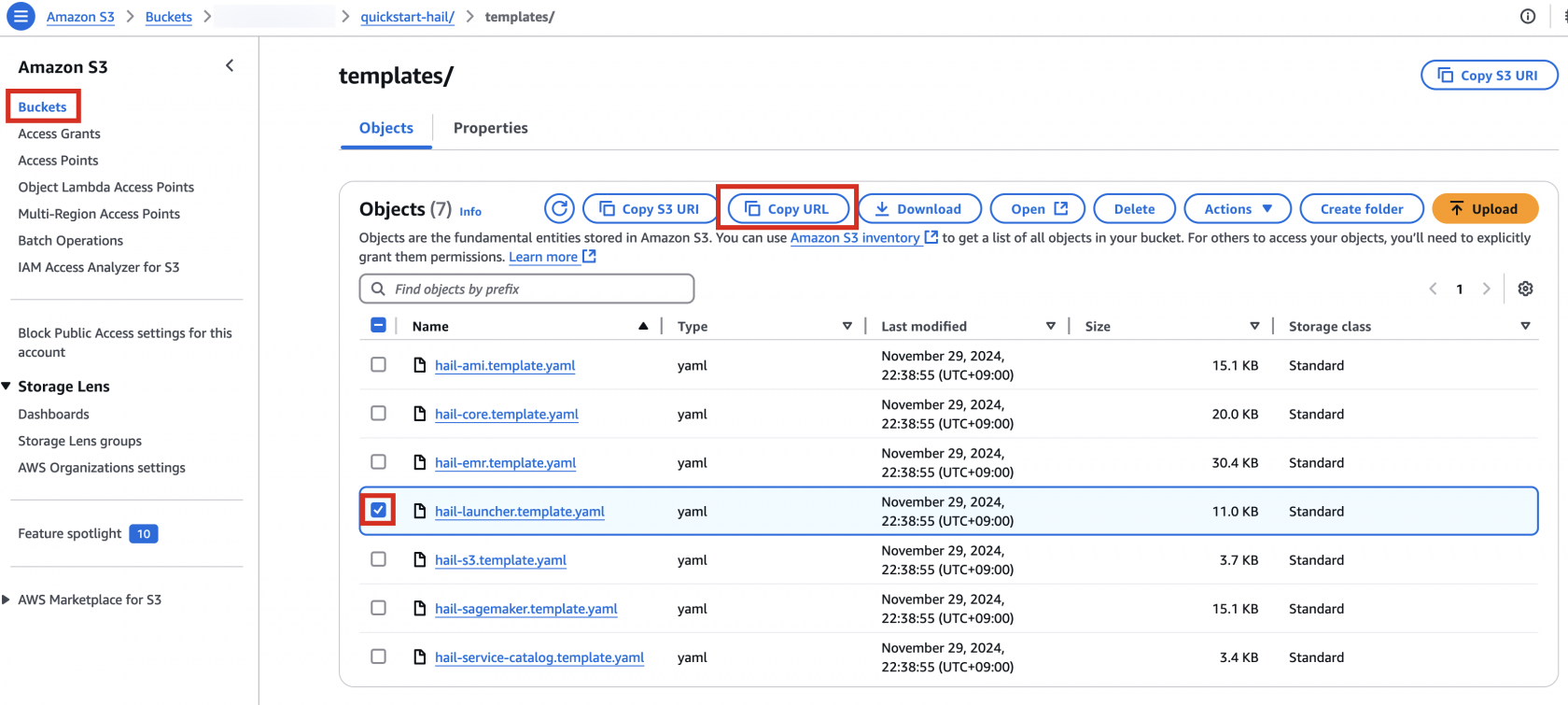
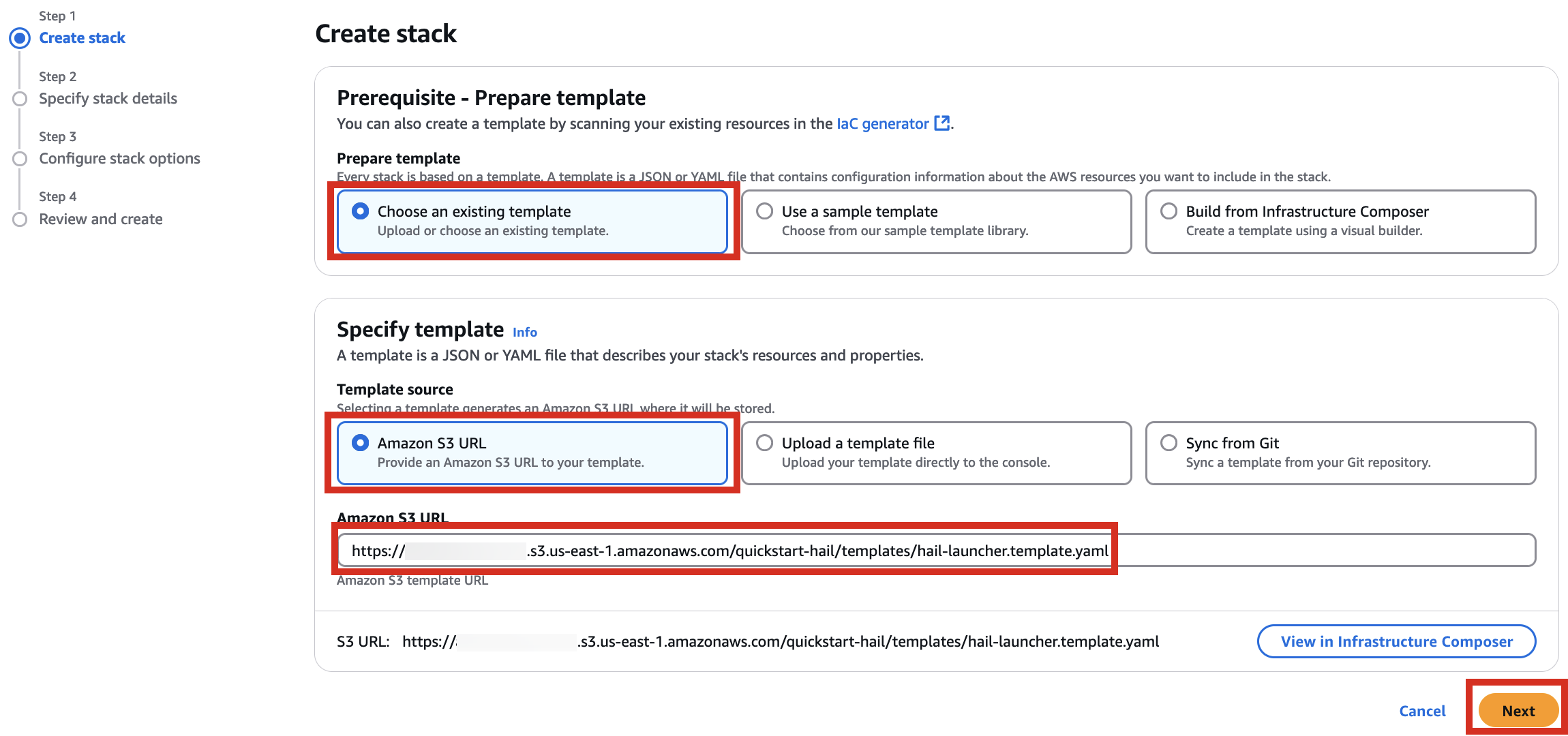
3. Go to the Amazon S3 console, select hail-launcher.template.yaml in the template directory you uploaded earlier, and click Copy URL. The path is as follows:
{bucket name} > quickstart-hail > templates > hail-launcher.template.yaml
When creating a CloudFormation stack, enter this URL and create the stack.
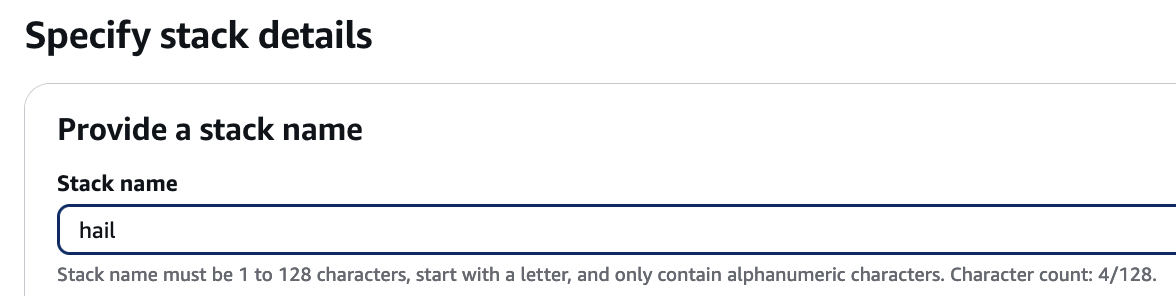
4. Proceed with entering information to create a stack.
Type an name for the stack.
Select a VPC. Select one subnet within the same VPC. For this exercise, select public.

Let's set it up to create additional buckets as needed.
Enter the name of the existing bucket where the quickstart-hail folder was uploaded, and check the region.
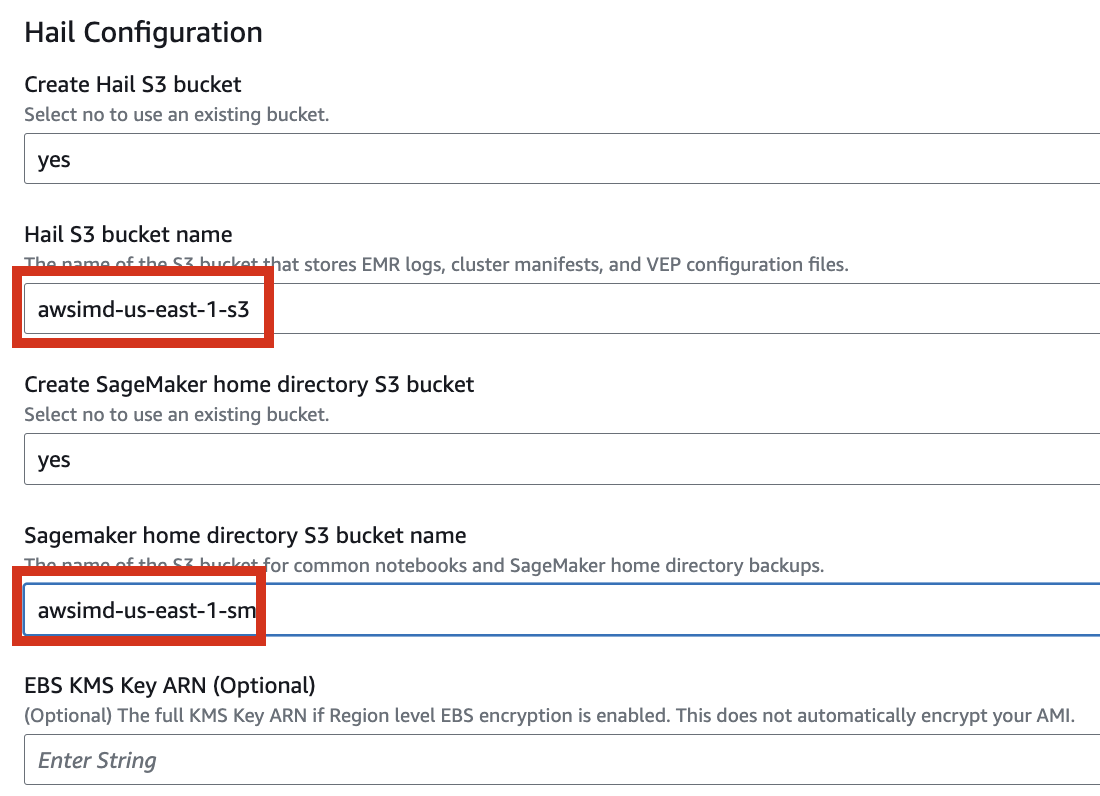
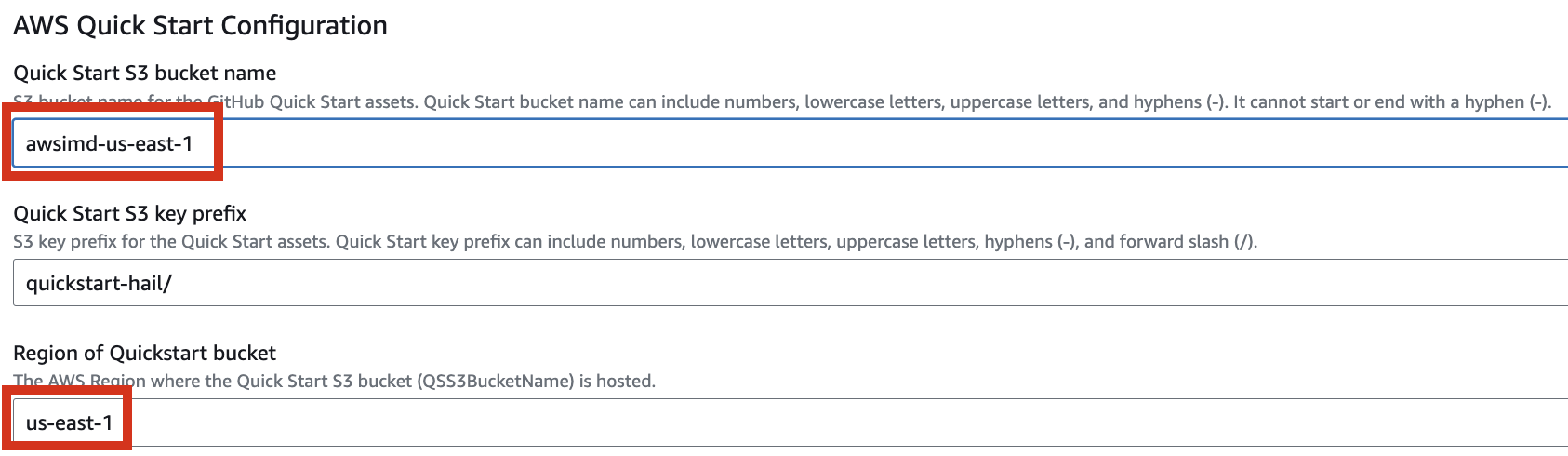
5. Finally, press the Next button to create the stack.
![]()
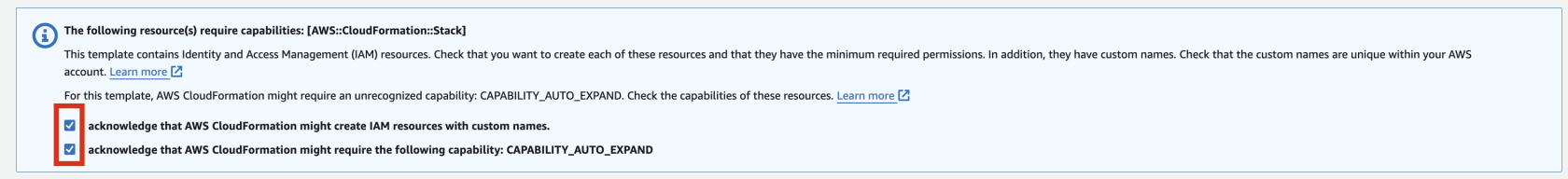
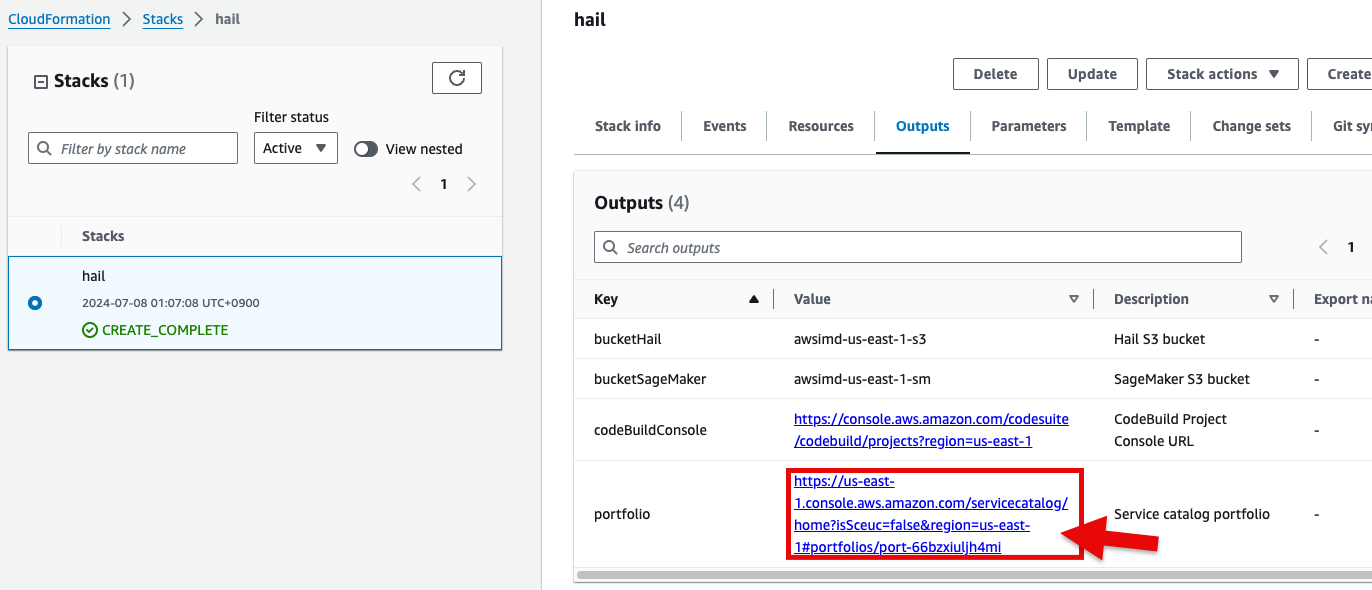
6. Check stack creation in CloudFormation.
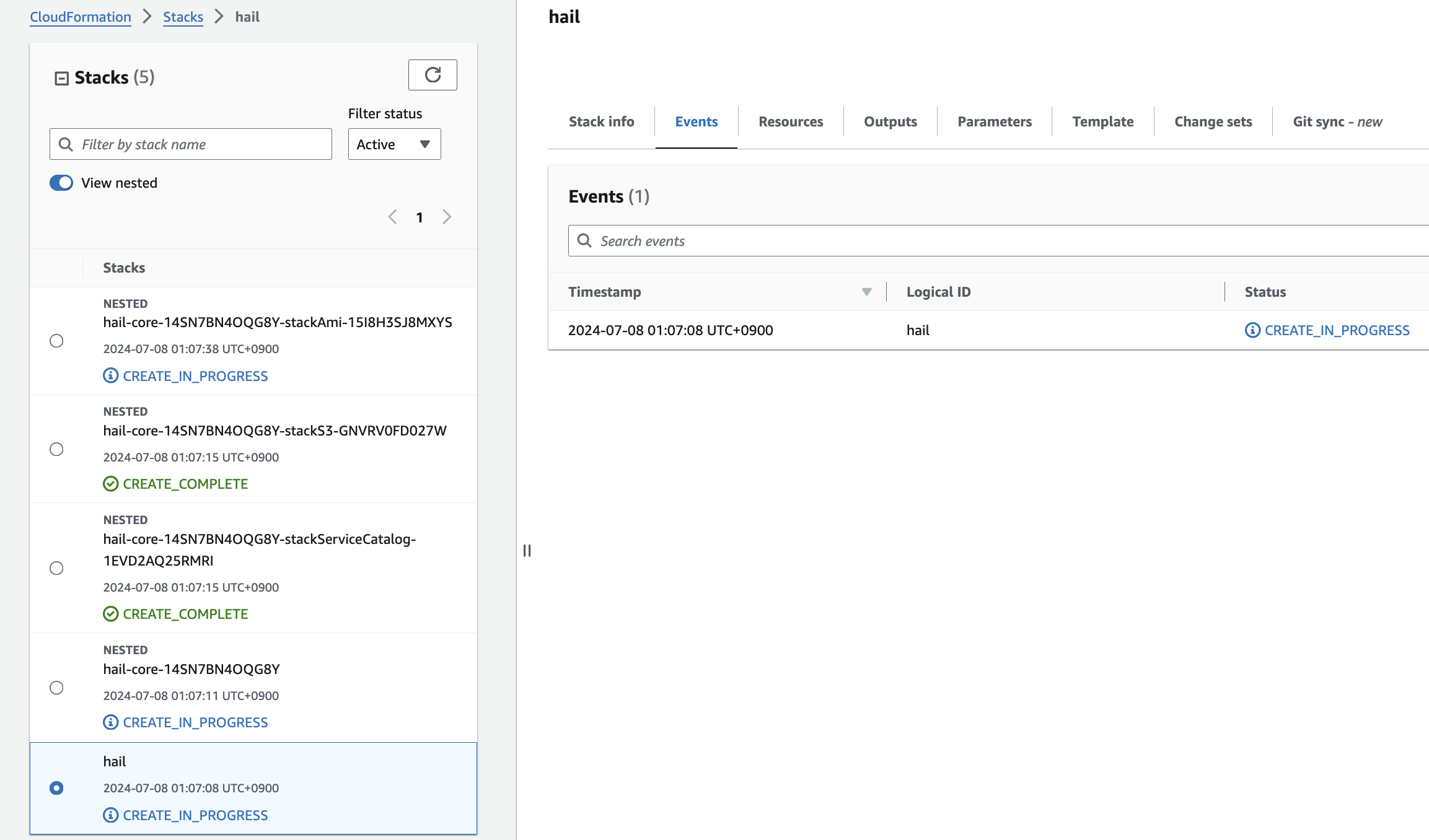
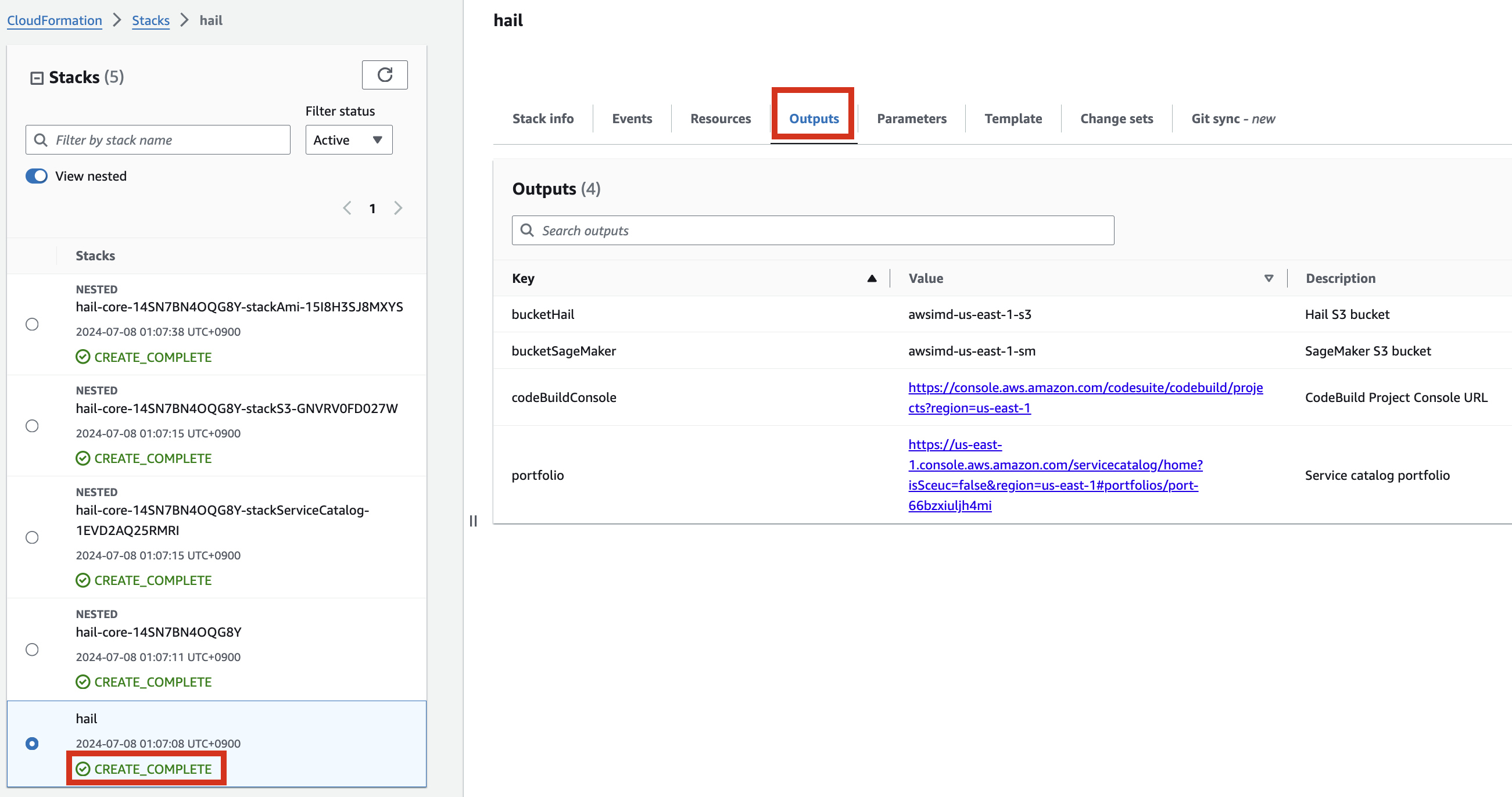
If the following portfolio appears in the output along with the CREATE_COMPLETE message in the top stack, you can confirm that it was executed correctly.
Create an AMI for Hail and VEP
Pre-downloading VEP Data and Storing in Bucket
For VEP, you can pre-download the data and store it in the bucket created or specified through the stack (using the bucketHail value from CloudFormation's Outputs).
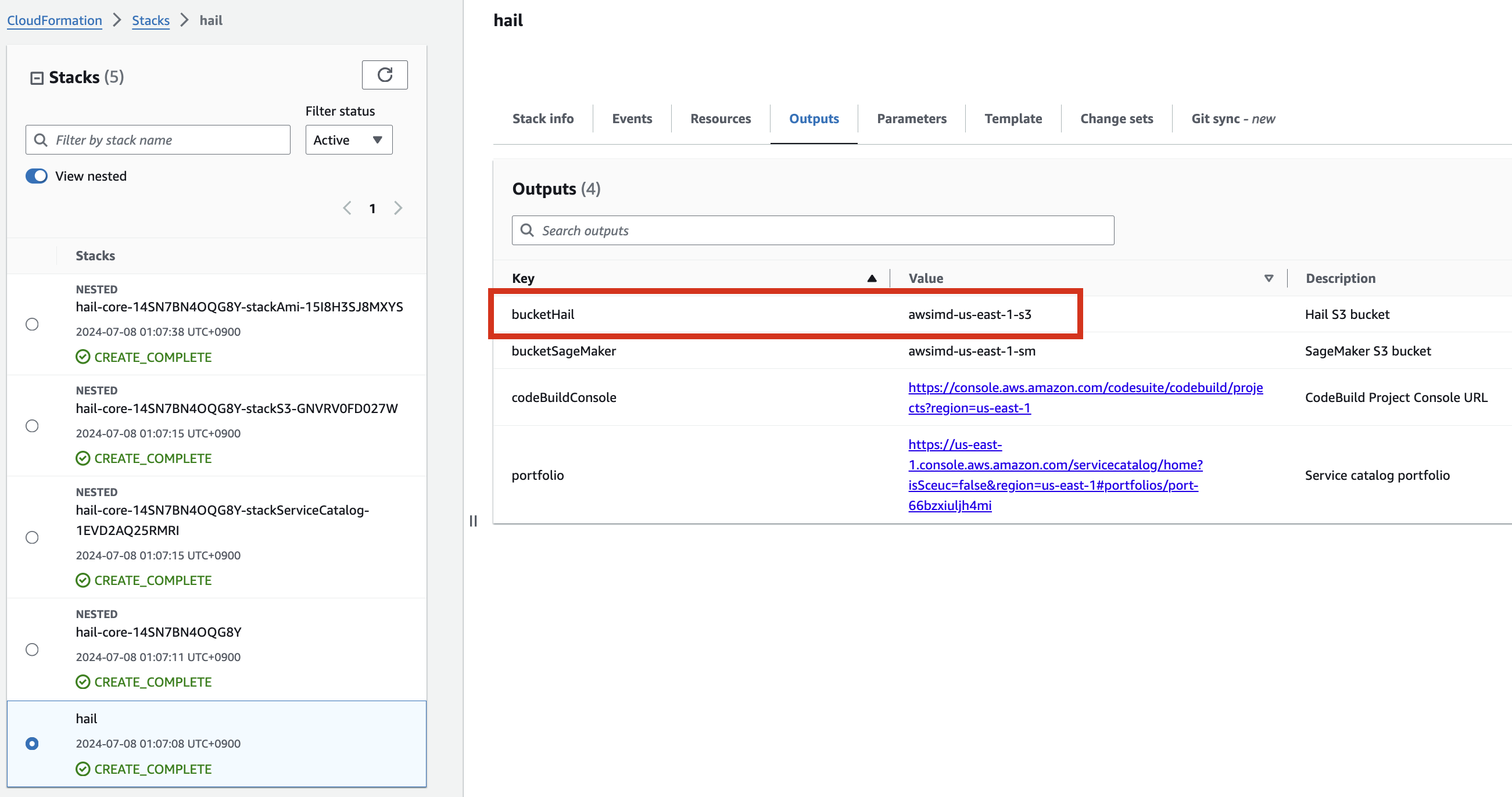
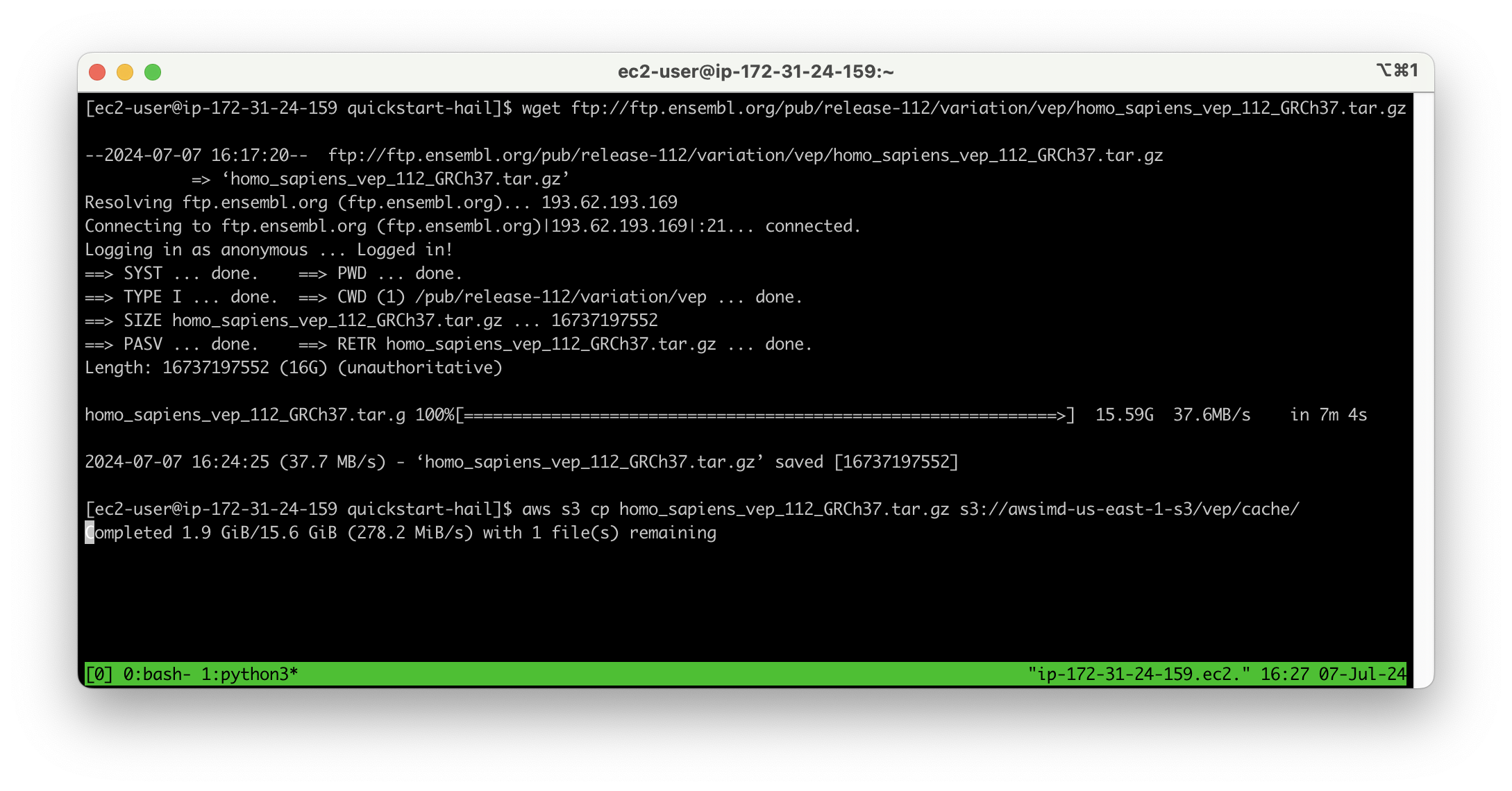
Download VEP data using the wget command:
wget ftp://ftp.ensembl.org/pub/release-112/variation/vep/homo_sapiens_vep_112_GRCh37.tar.gzUpload the downloaded file to your bucket: (I emphasize that this is the value for the BucketHail key confirmed in CloudFormation Outputs)
aws s3 cp homo_sapiens_vep_112_GRCh37.tar.gz s3://{bucketdefined name}Hail S3 bucket}/vep/cache/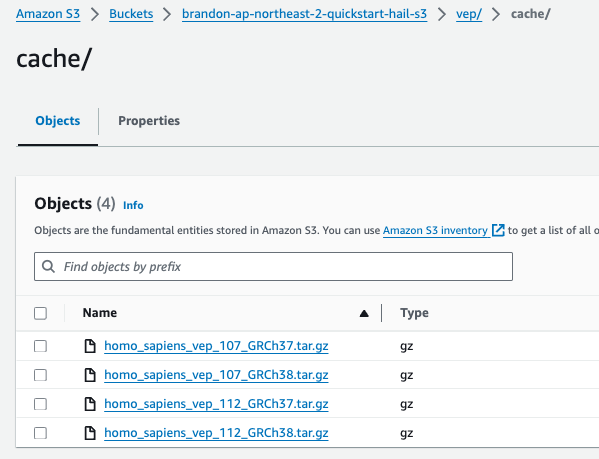
AMI Build
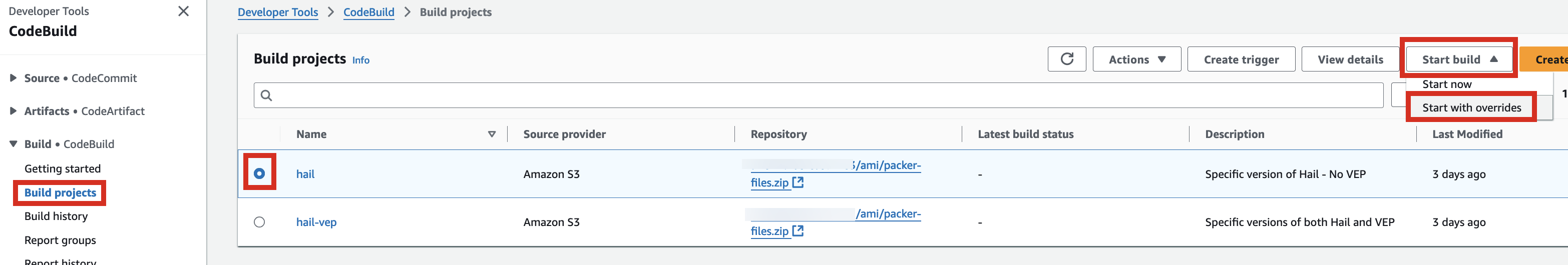
1. Access the CodeBuild console and initiate the build process for each new AMI. Select Start build > Start with overrides.
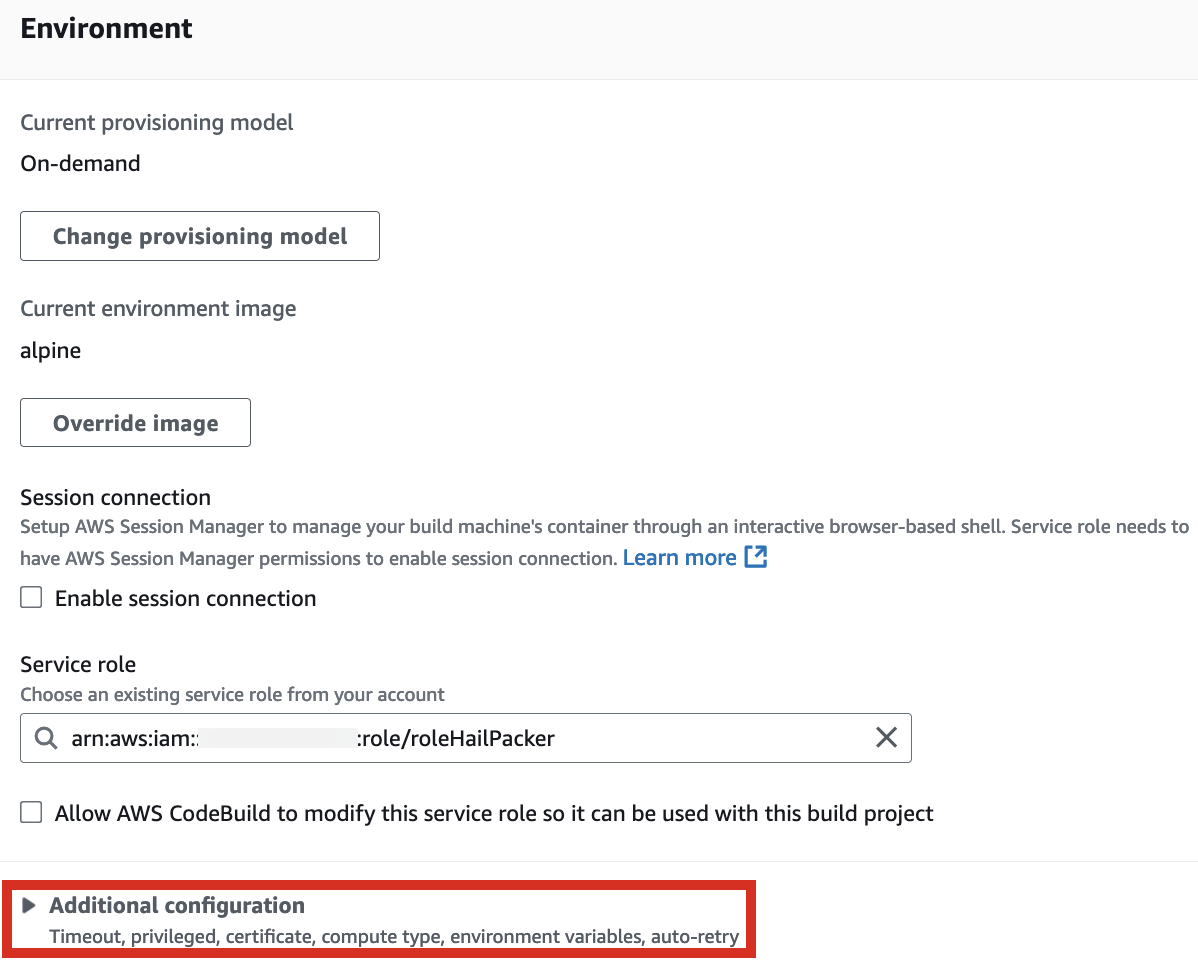
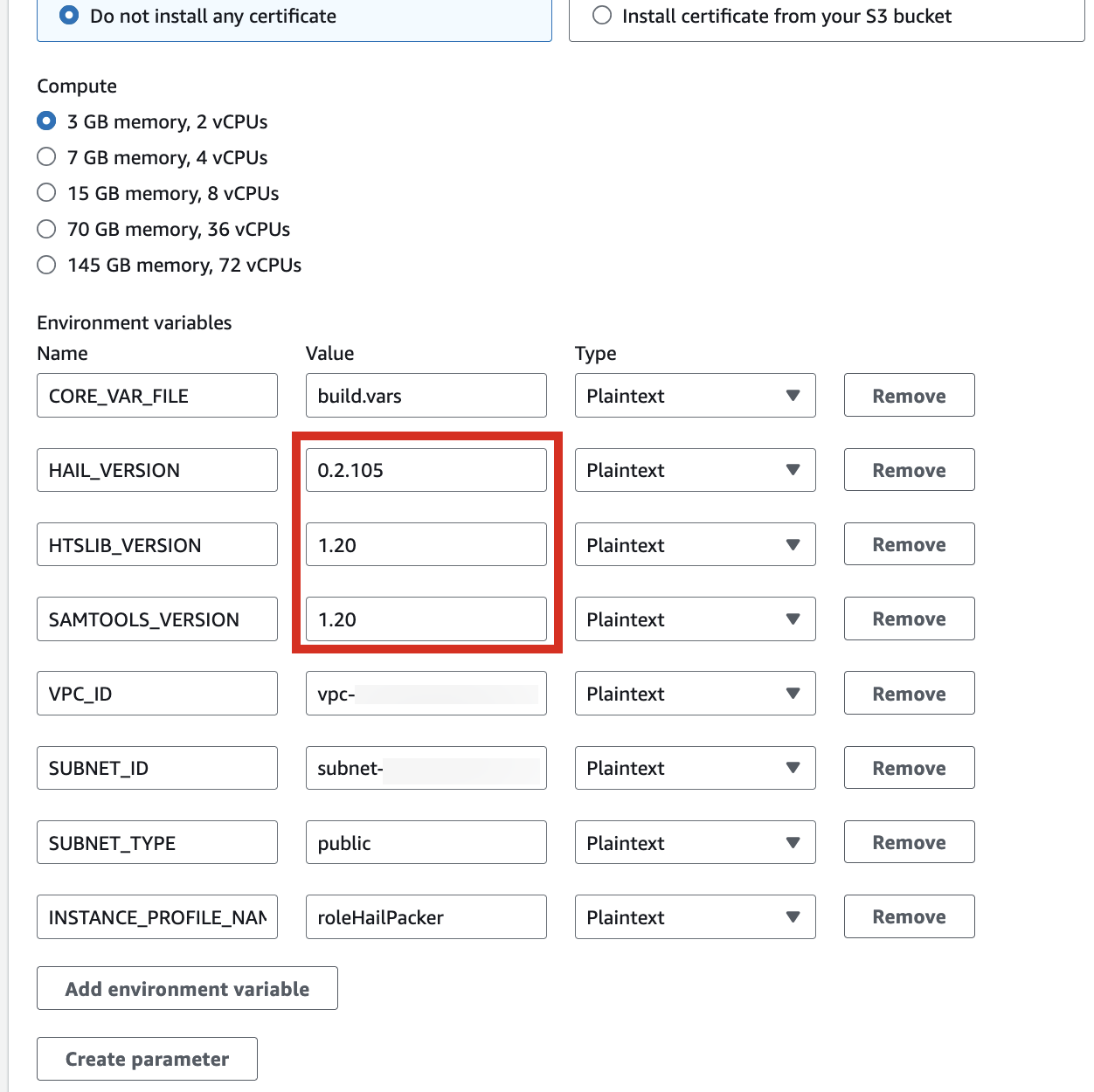
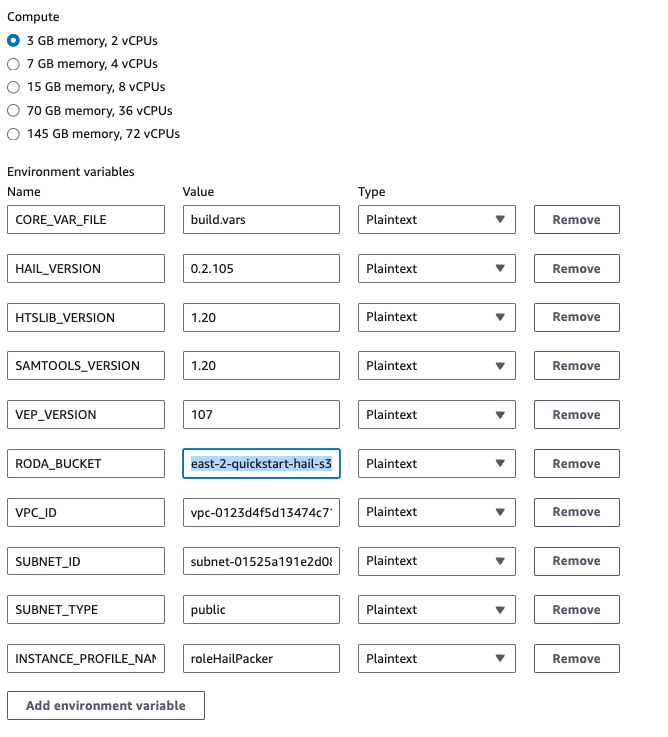
2. In the Environment section, expand Additional configuration and input the required values.
| HAIL_VERSION | 0.2.105 |
|---|---|
| HTSLIB_VERSION | 1.20 |
| SAMTOOLS_VERSION | 1.20 |
* For building with hail-vep option (includes VEP installation):
| HAIL_VERSION | 0.2.105 |
|---|---|
| HTSLIB_VERSION | 1.20 |
| SAMTOOLS_VERSION | 1.20 |
| VEP_VERSION | |
| RODA_BUCKET | <VEP download bucket name> |
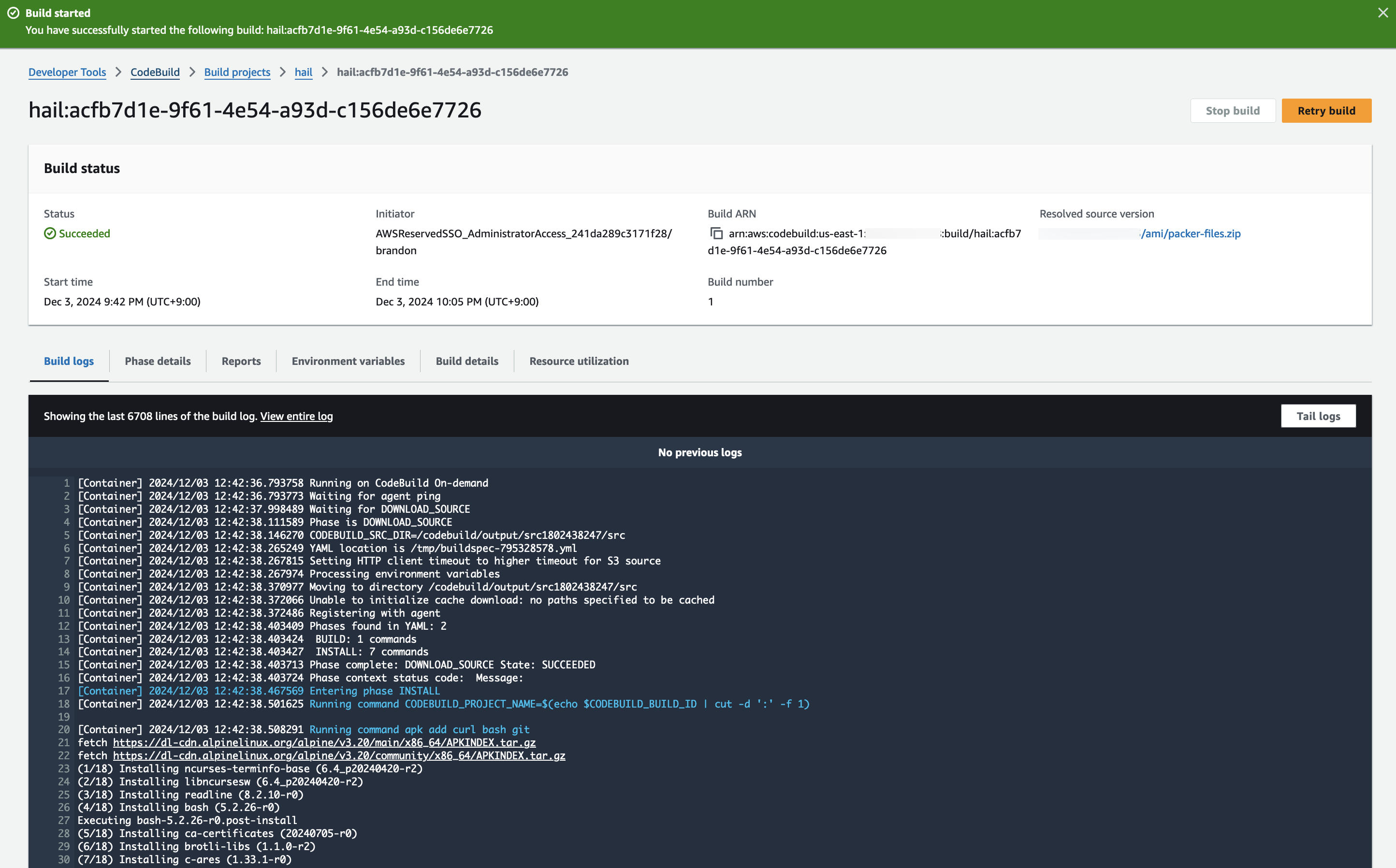
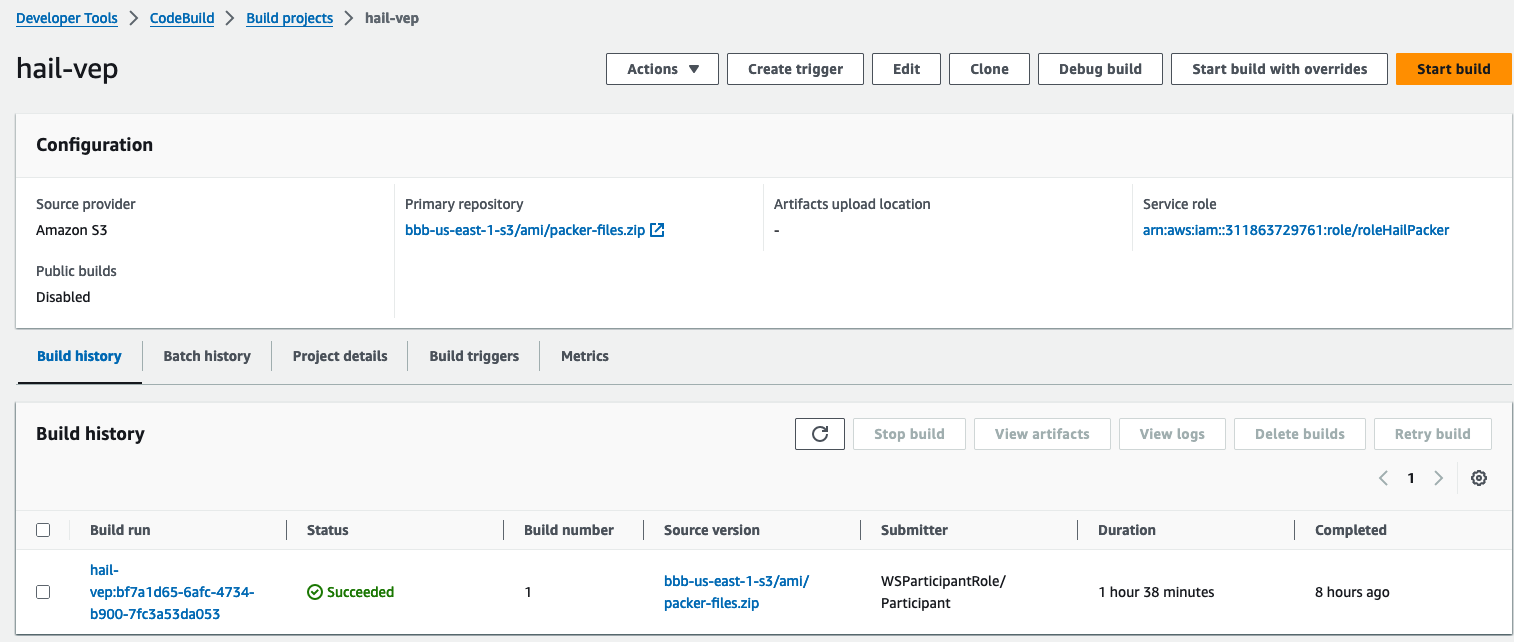
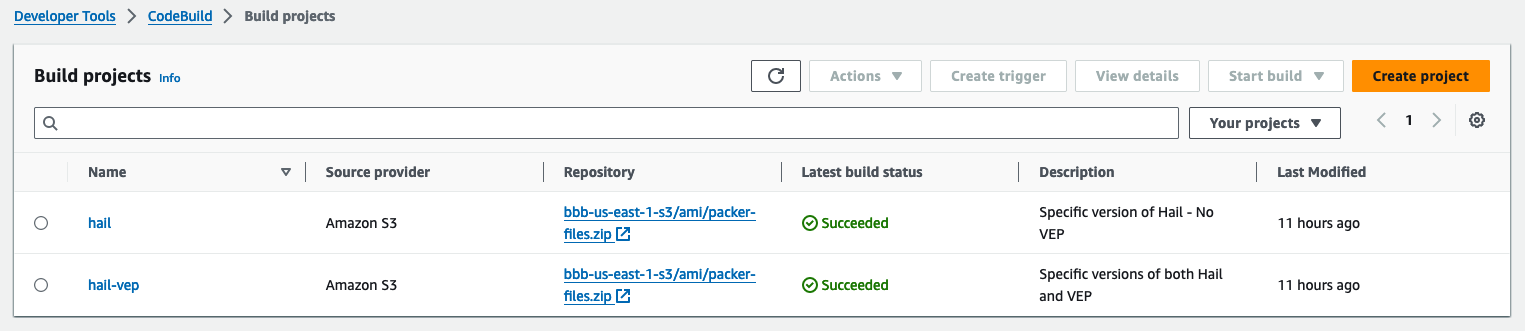
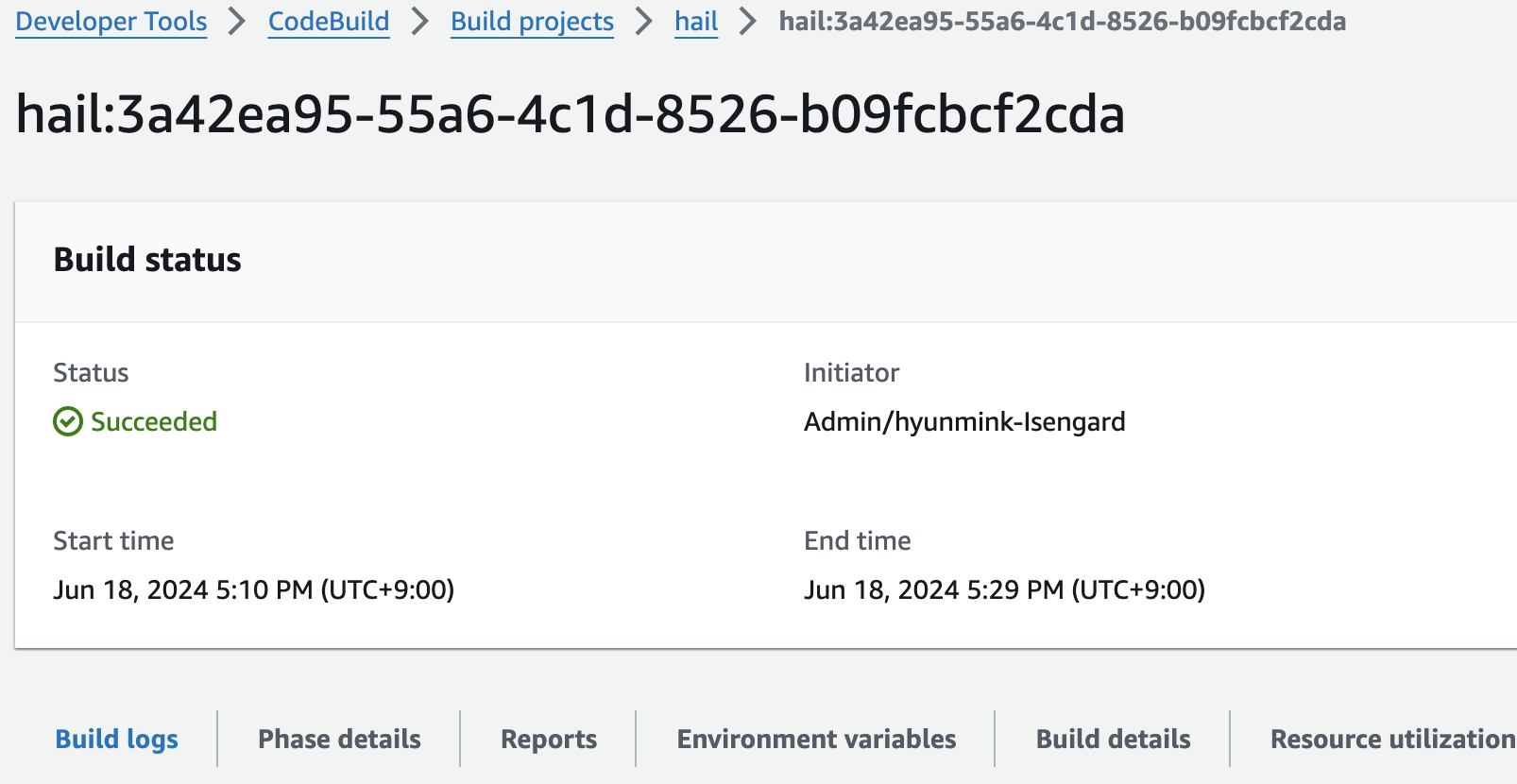
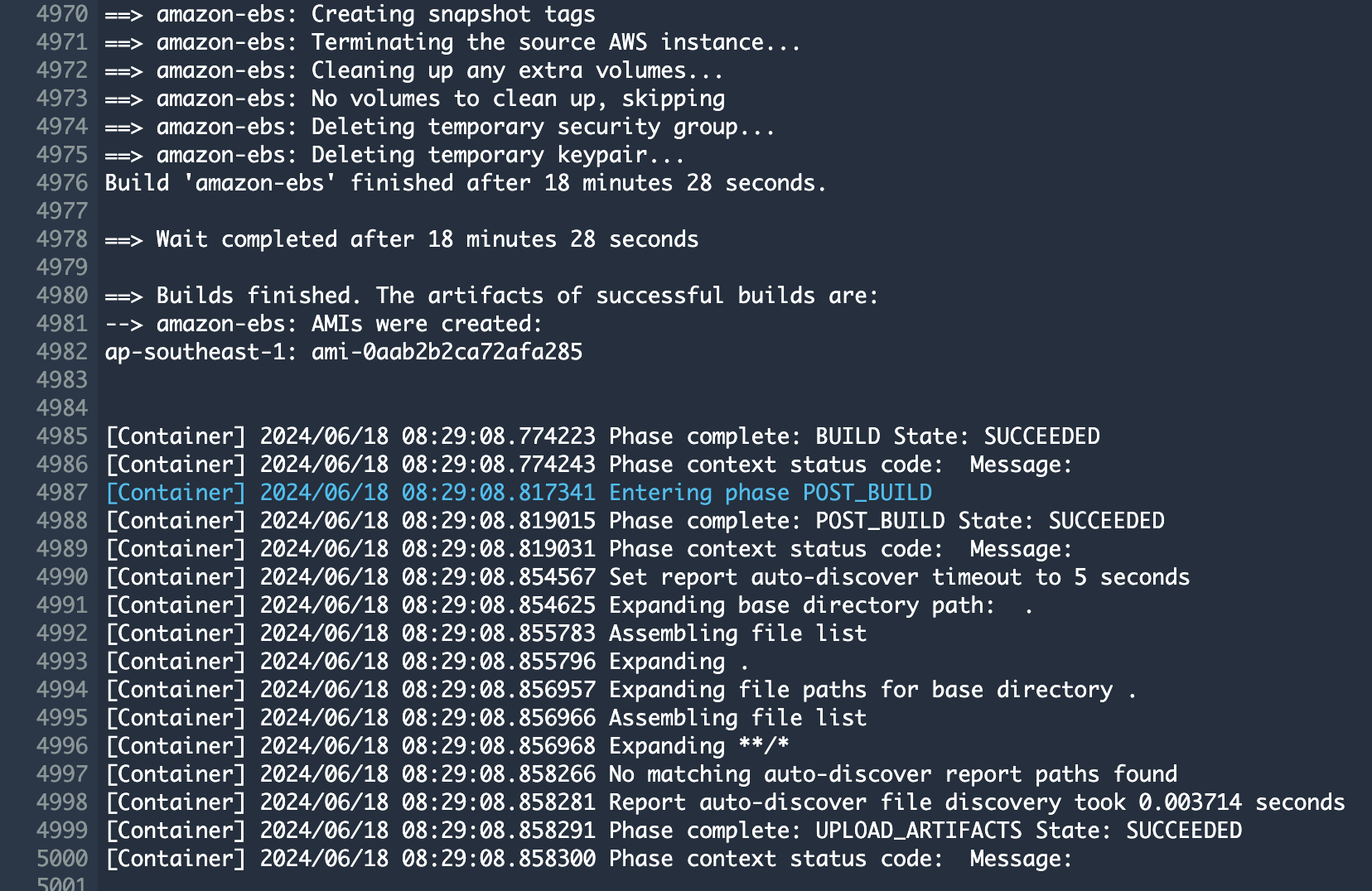
3. Check the build status
Hail (without VEP): The Hail image build completes in about 20 minutes.
Hail (VEP): The VEP version build takes approximately 1 hour and 38 minutes to complete.
The Hail image build completes in about 20 minutes.
You can find the **AMI results** in either the [AMI menu of Amazon EC2 console or CodeBuild logs.
또는
EMR 클러스터Cluster 실행Setup 및and Jupyter 환경Environment 세팅Configuration
EMR 클러스터Cluster 실행Setup
- In the CloudFormation
서비스service콘솔에서 스택console's Outputs탭의tab, click the portfolio에 있는 링크를 클릭합니다.
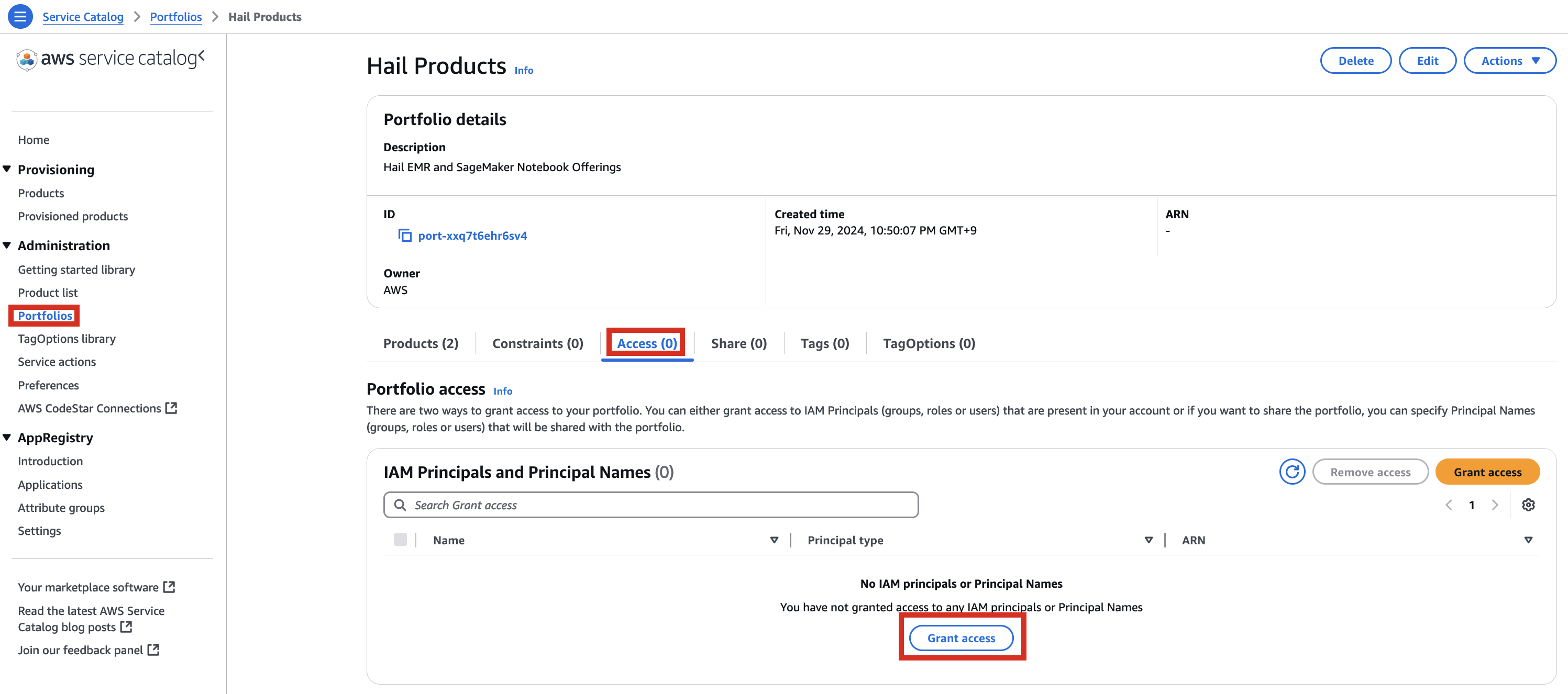
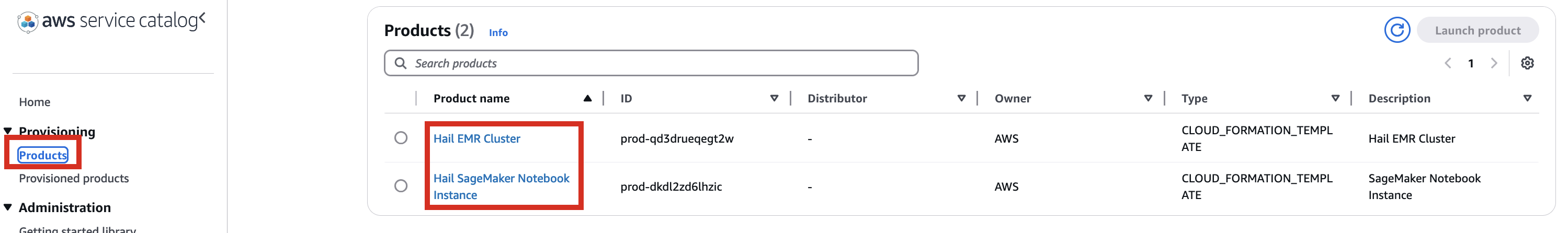
1.

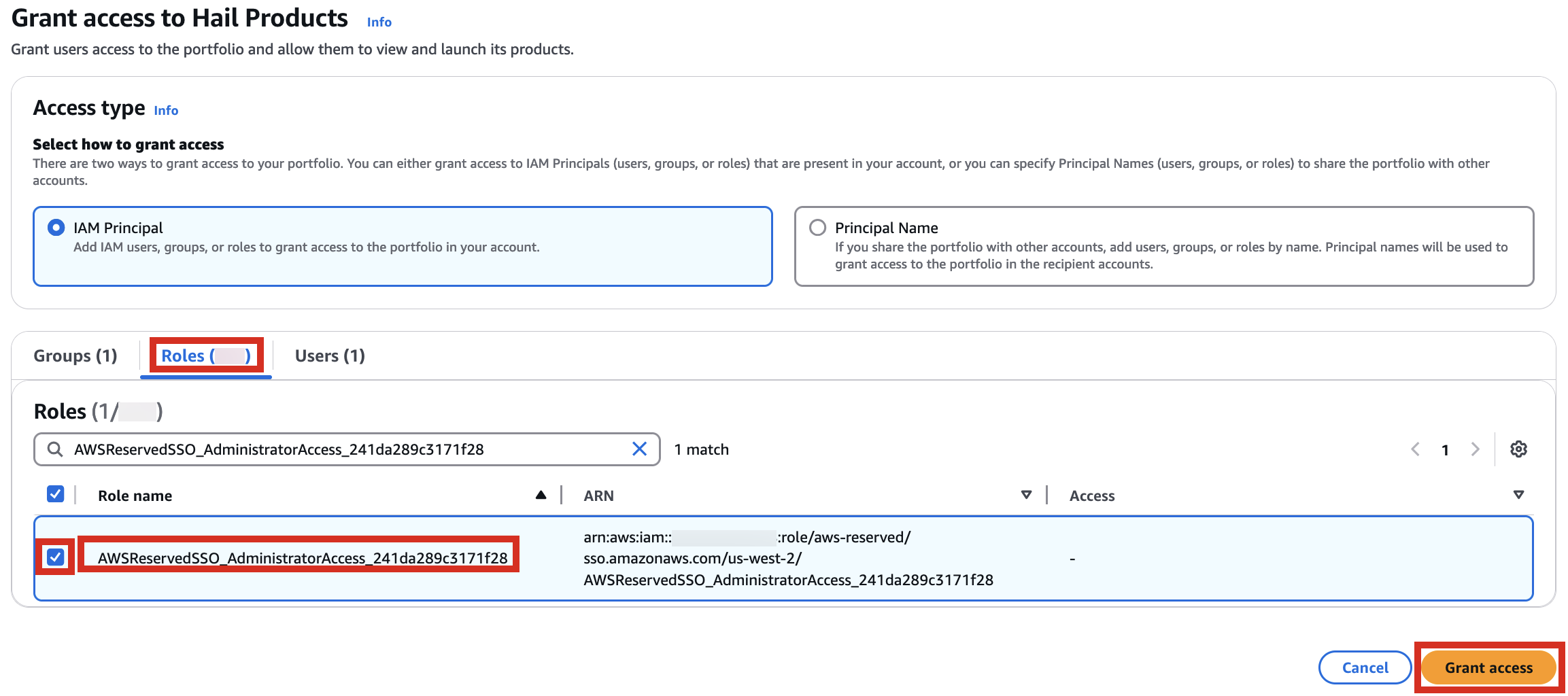
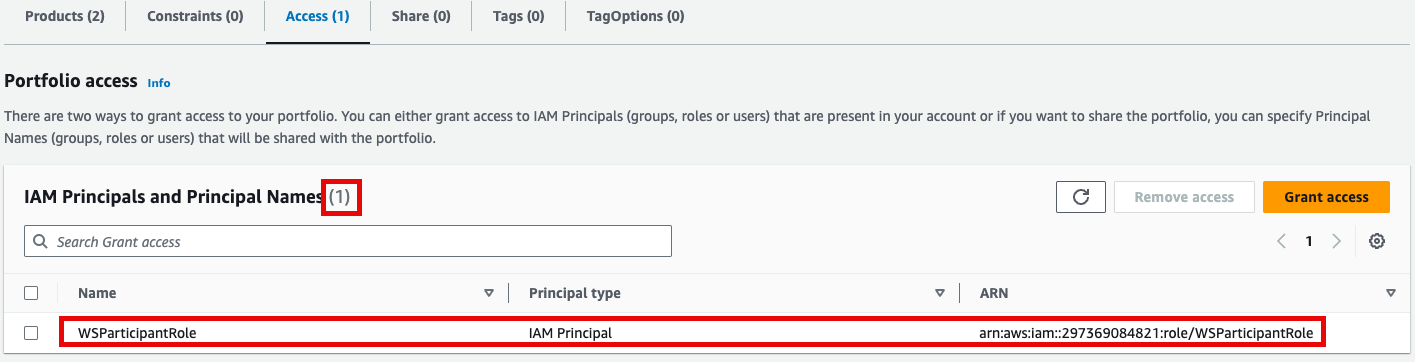
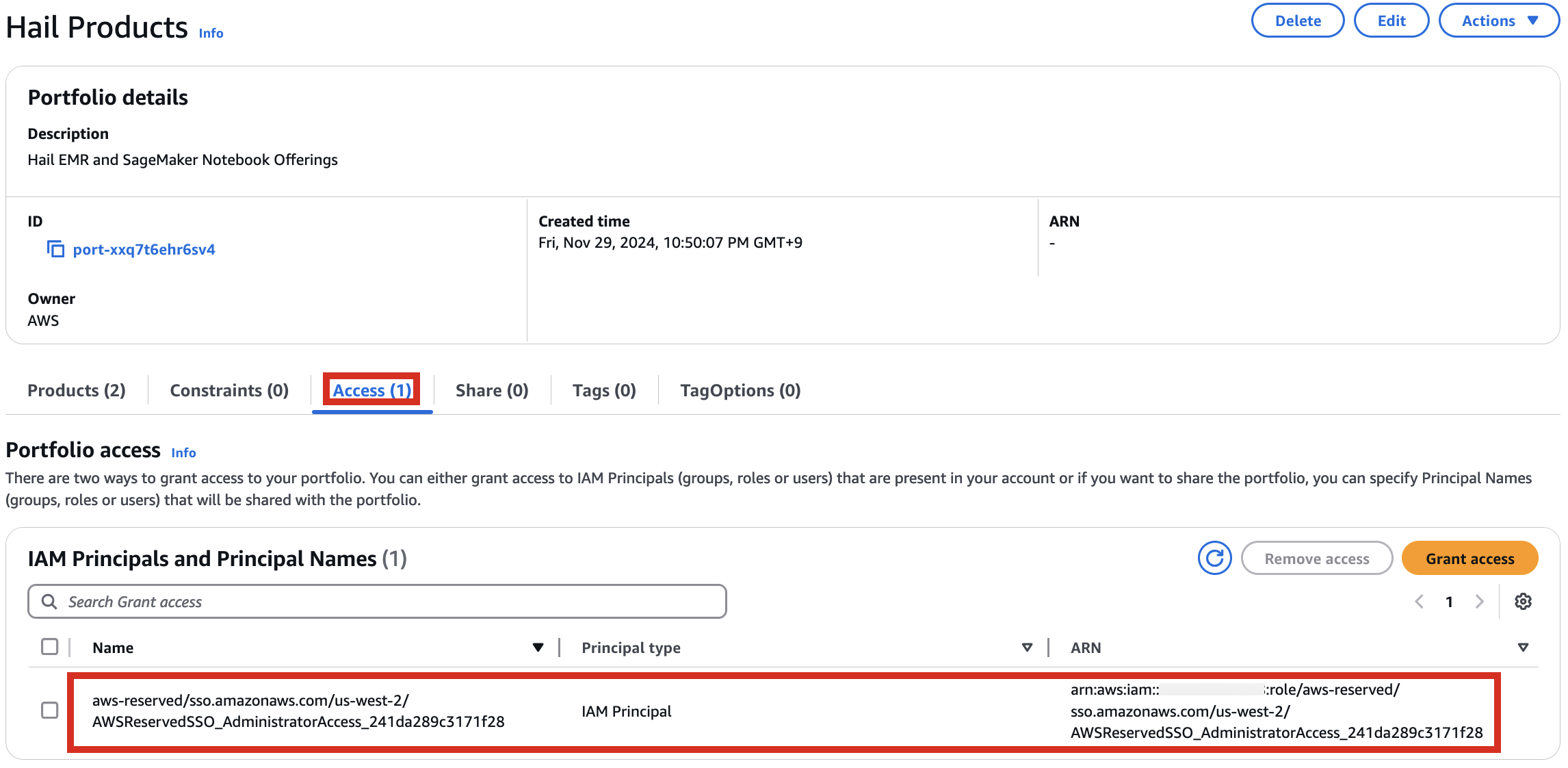
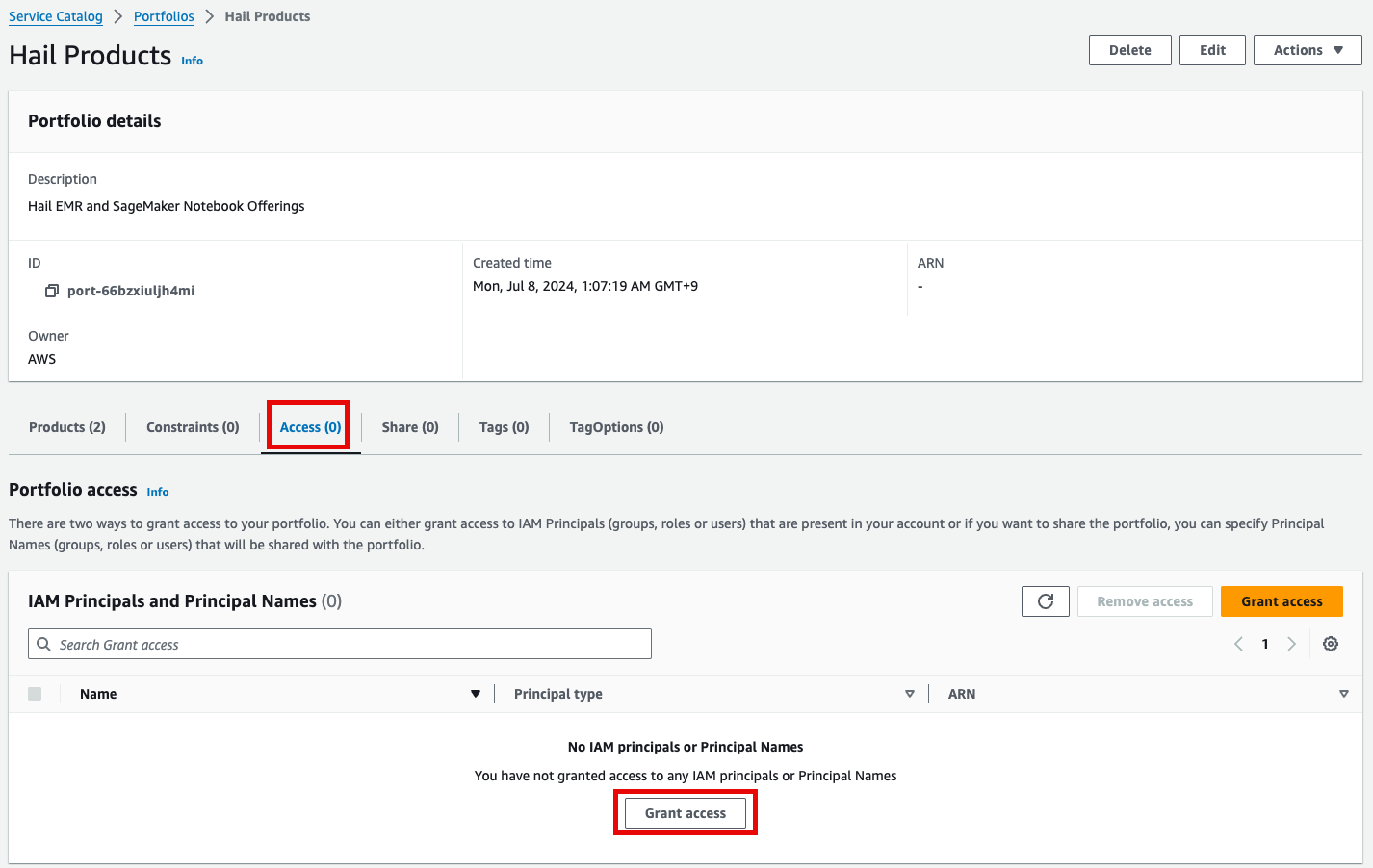
포트폴리오내In해당theProduct에portfolio,대한locate the relevant Product, click the Access탭을tab,클릭한then뒤select Grant access.
3. Add permissions. Check the user that suits you and grant them access
를to클릭합니다.Hail
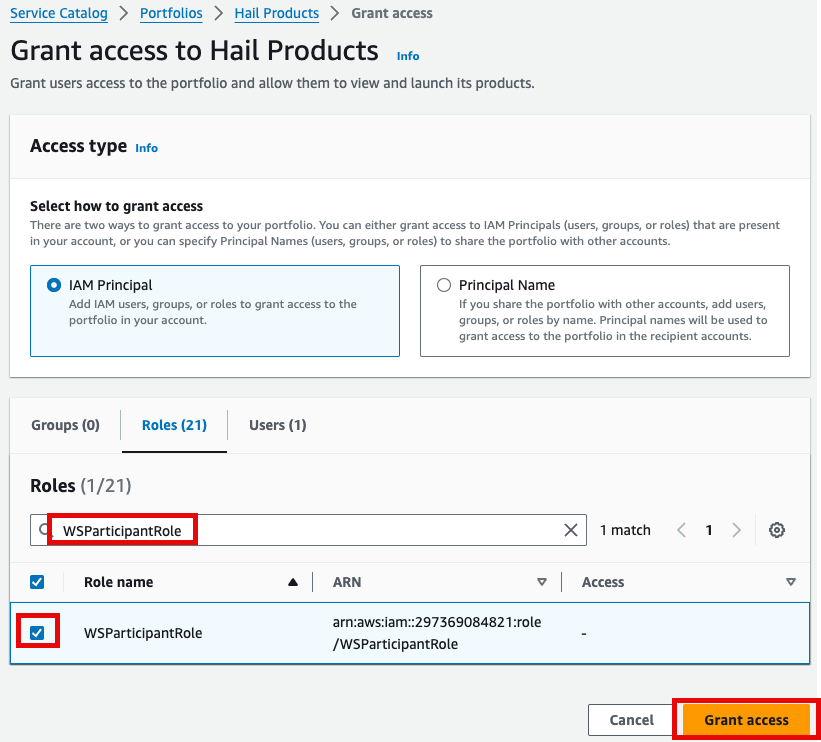
2.
- for
권한it,추가를select합니다.it,여기서는and실습 계정 역할 이름이WSParticipantRole입니다. 검색후 체크하고click Grantaccess를 클릭합니다.
Access4.
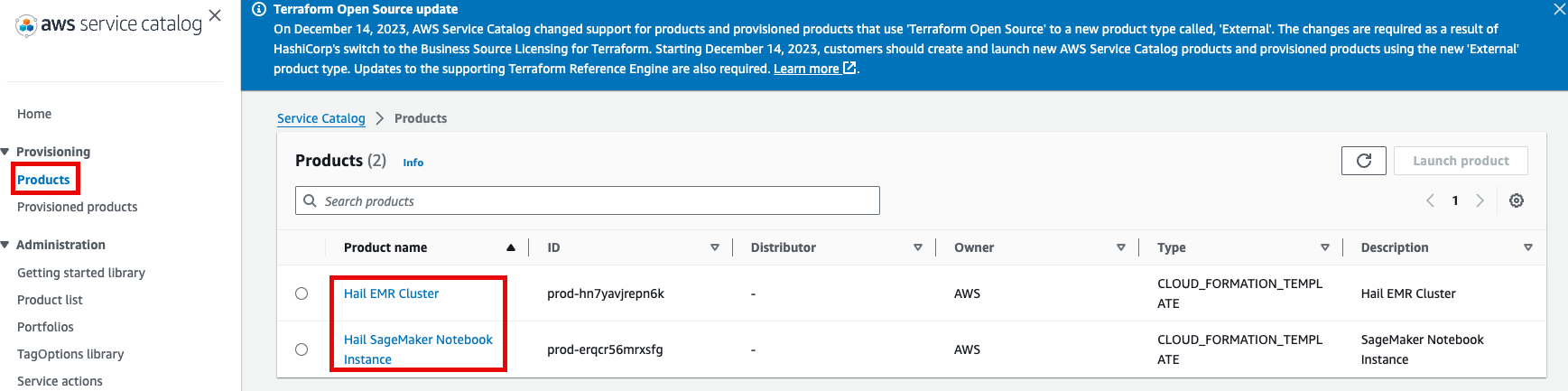
권한이After있음을confirming확인한access뒤permissions, navigate to the Product in the Provisioning메뉴의 Product를 클릭하여 진입합니다.
이제With권한이permissions있으므로granted, you should now see 2 Products항목에서 2개의 Product들을 볼 수 있게 되었습니다.
5.
Product에Select있는the Hail EMRCluster메뉴로Cluster진입하여product원하는andproduct를 선택하고click Launchproduct를 클릭합니다.
6.
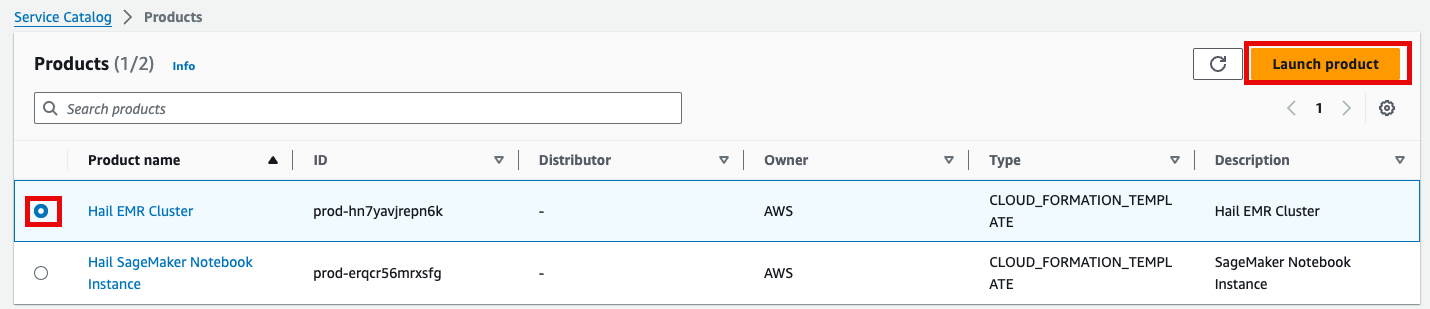
Launch에Enter필요한the정보들을required기입합니다.launch
7.
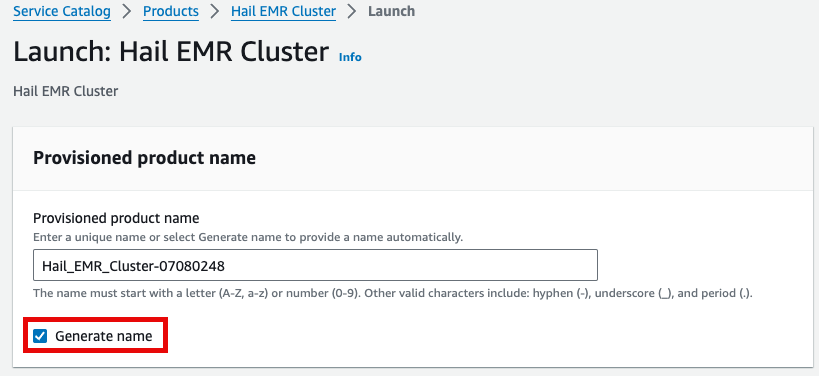
이름을Either 직접enter 입력하거나a name manually or click Generate name을 클릭합니다.name.
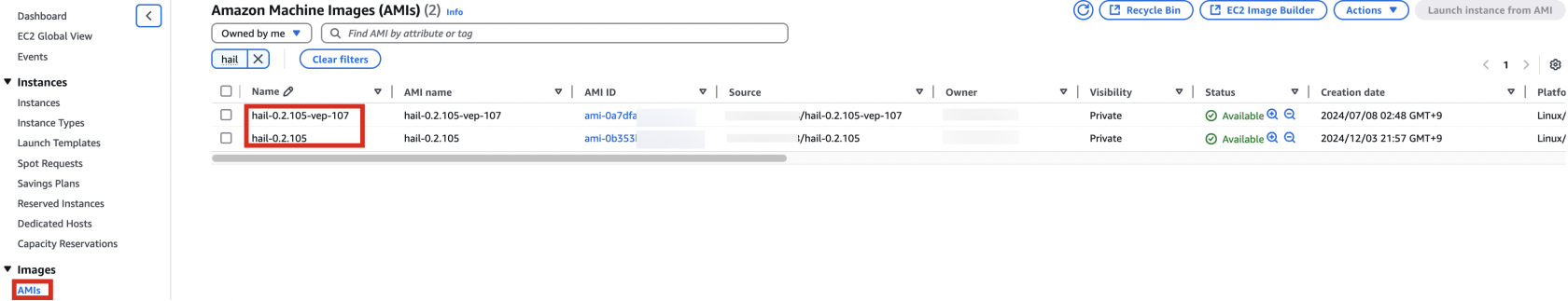
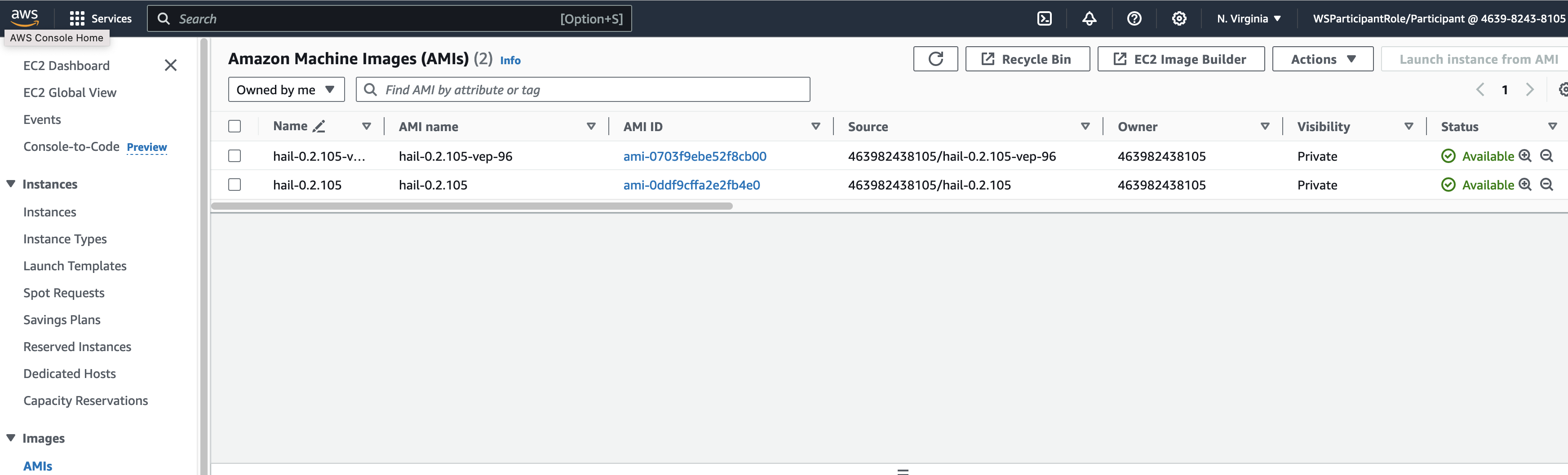
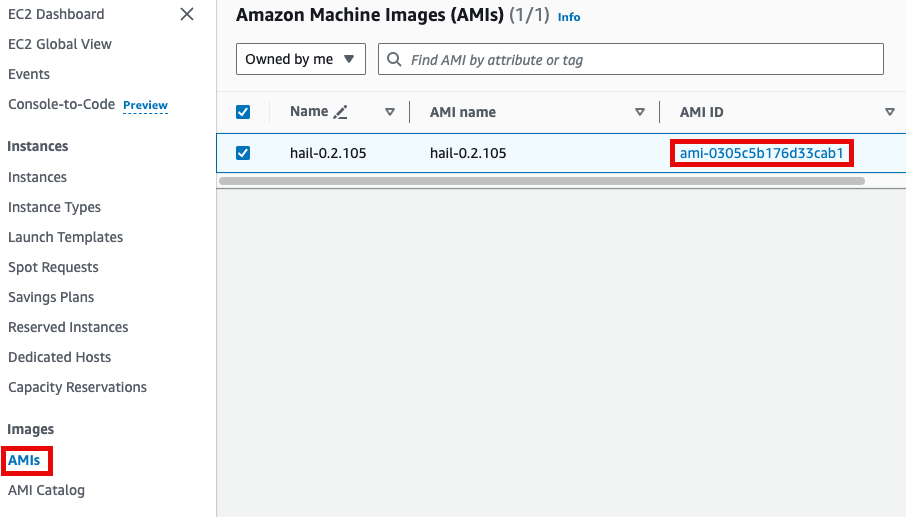
앞에서Specify 만든the Hail AMI를AMI 입력합니다.you 이때created earlier. You can find the AMI ID는ID AMIin 메뉴에서the EC2 서비스 하위의service's AMIs 항목에서 찾을 수 있습니다.section (앞에서도as 설명했던)previously described).
Input the Cluster name을name 입력하고and Hail AMI에 AMI ID를ID. 입력한You 뒤can 다른leave 것은all 모두other 기본값을settings 사용할at 수their 있습니다.default values.
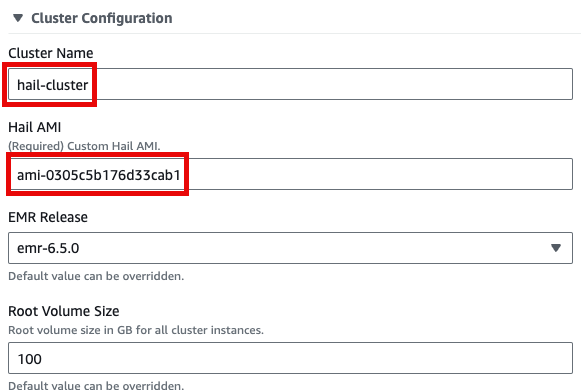
맨 아래의Click Launchproduct를product클릭합니다.at
8.

SageMaker Notebook 실행
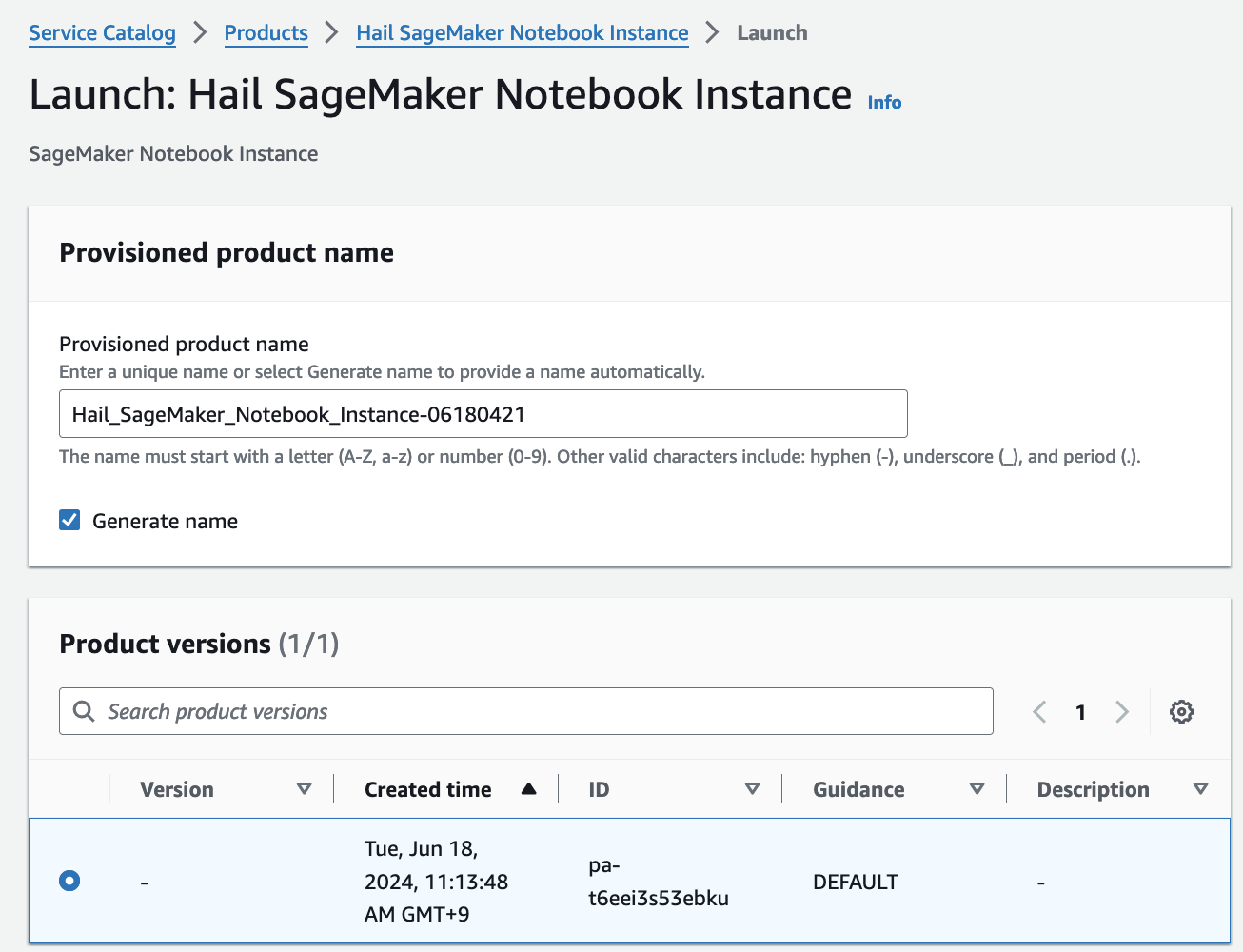
- Product 메뉴에서 마찬가지로 Launch product를 클릭합니다.
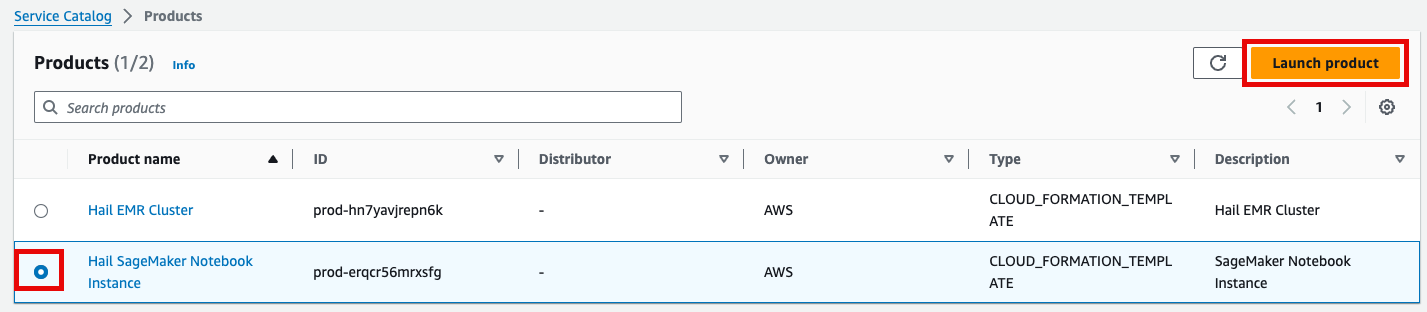
- Hail notebook을 위한 인스턴스의 이름을 입력하고 나머지는 모두 기본값입니다.
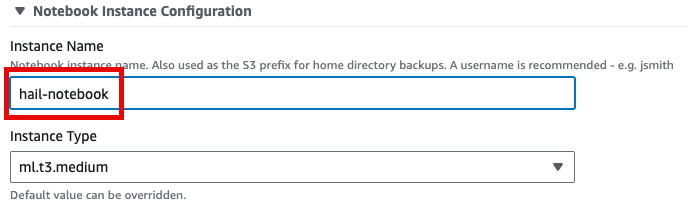
- 맨 아래의 Launch product를 클릭합니다.

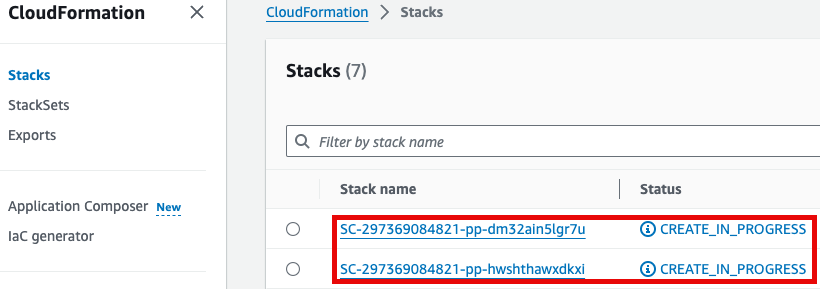
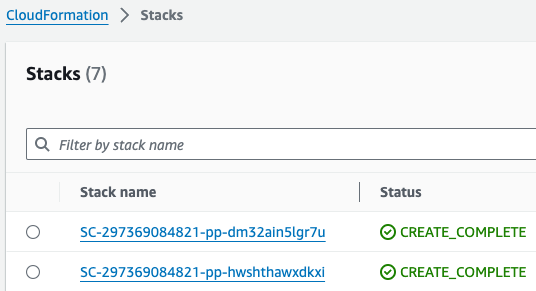
참고로 제품(Product)의 실행 과정은 CloudFormation을 통해서도 확인할 수 있습니다.
GWAS 실습 (Hail)
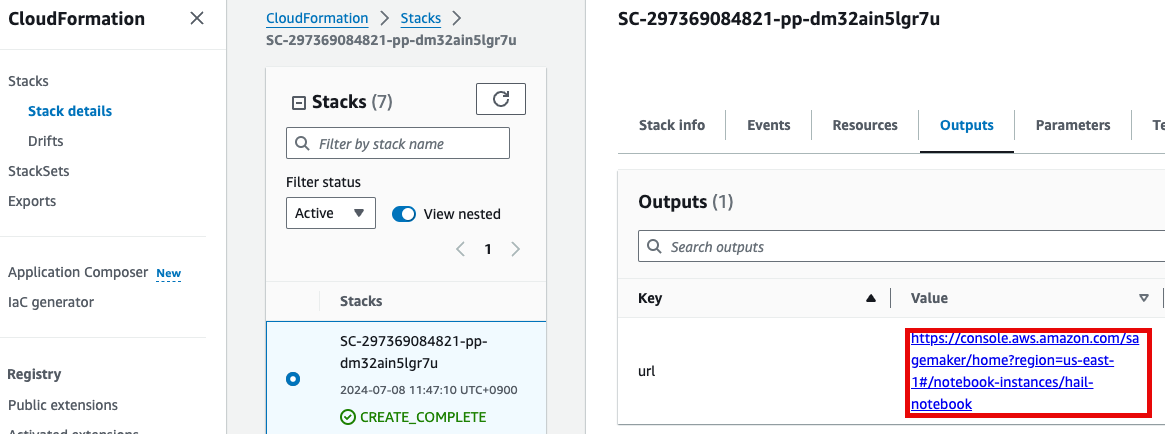
- 노트북을 실행합니다. 이 때 CloudFormation의 Outputs탭에서 url을 확인할 수 있습니다. 클릭하면 Amazon SageMaker의 해당 노트북 인스턴스로 자동 연결됩니다.
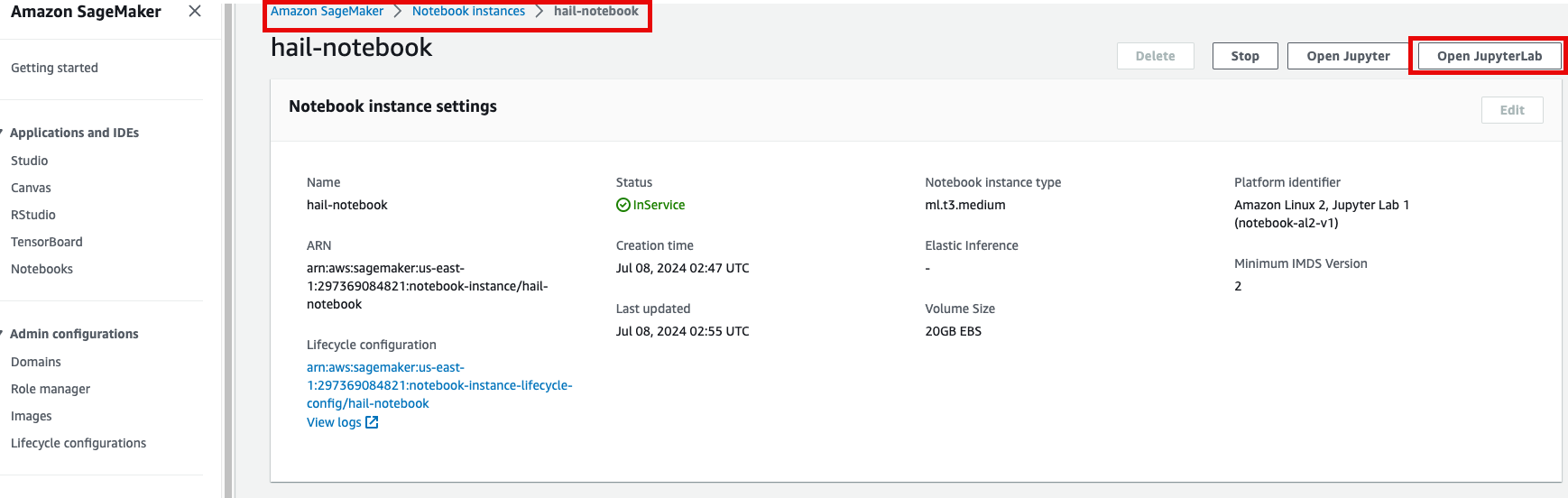
- Open JupyterLab을 클릭하여 노트북을 실행합니다.
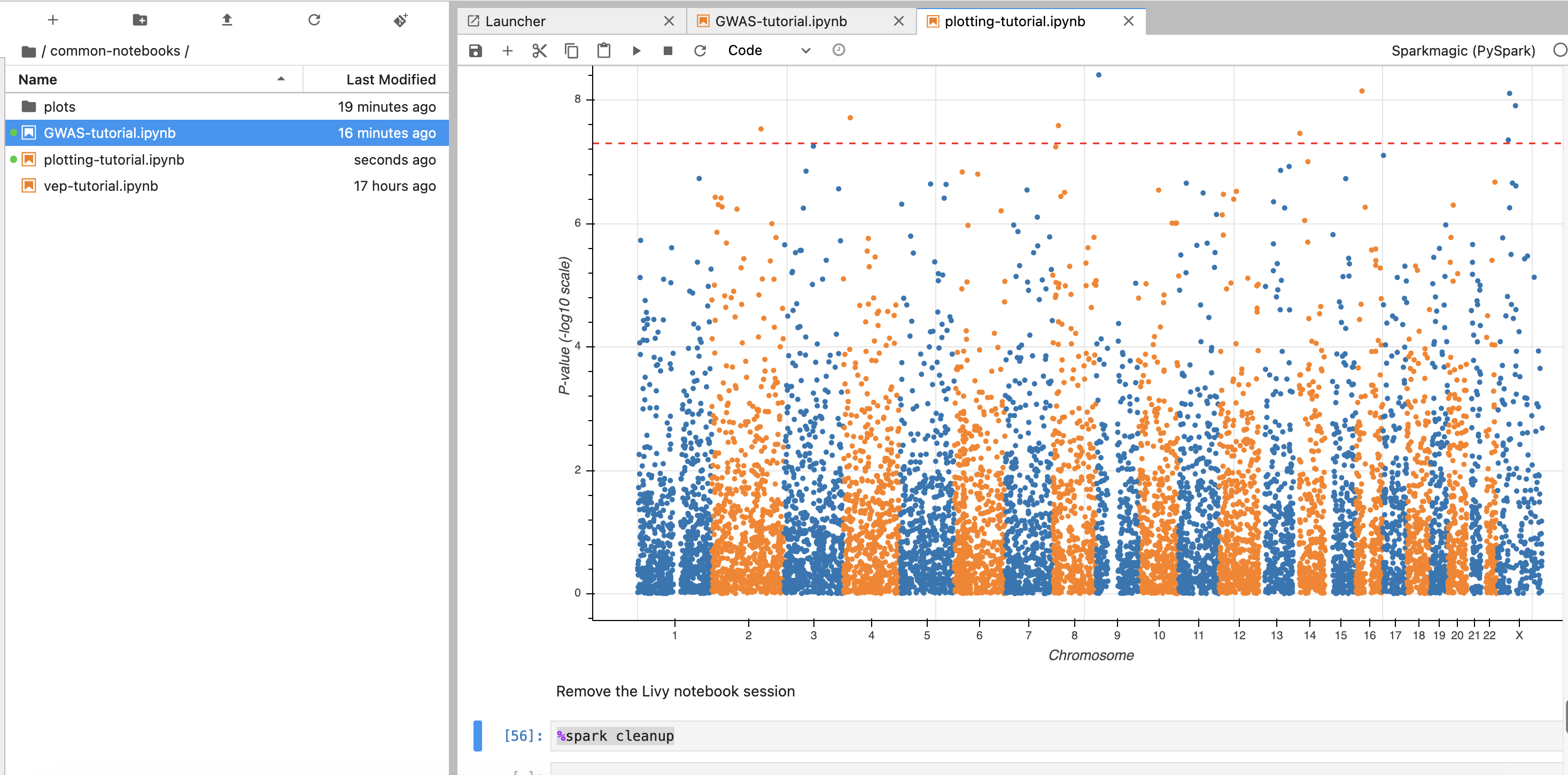
- 이제 노트북에서 각각 2개의 노트북을 가지고 실습해봅니다.
- common-notebooks/plotting-tutorail.ipynb
- common-notebooks/GWAS-tutorial.ipynb
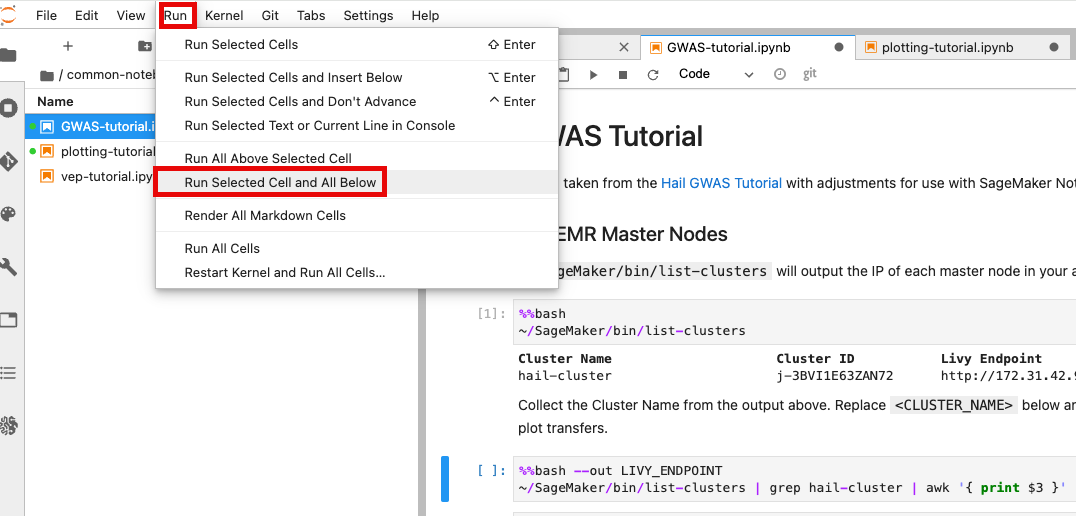
앞에서 만든 EMR 클러스터를 조회한 뒤 Cluster Name을 2번째 셀에서 수정해줍니다.
노트북 셀을 한번에 실행하기 위해서 시작하고자 하는 셀에 커서를 놓은 뒤 일괄 실행할 수 있습니다.
최종적으로 튜토리얼로 주어진 코드가 정상적으로 실행할 수 있었다면 아래와 같은 결과들을 확인할 수 있습니다.
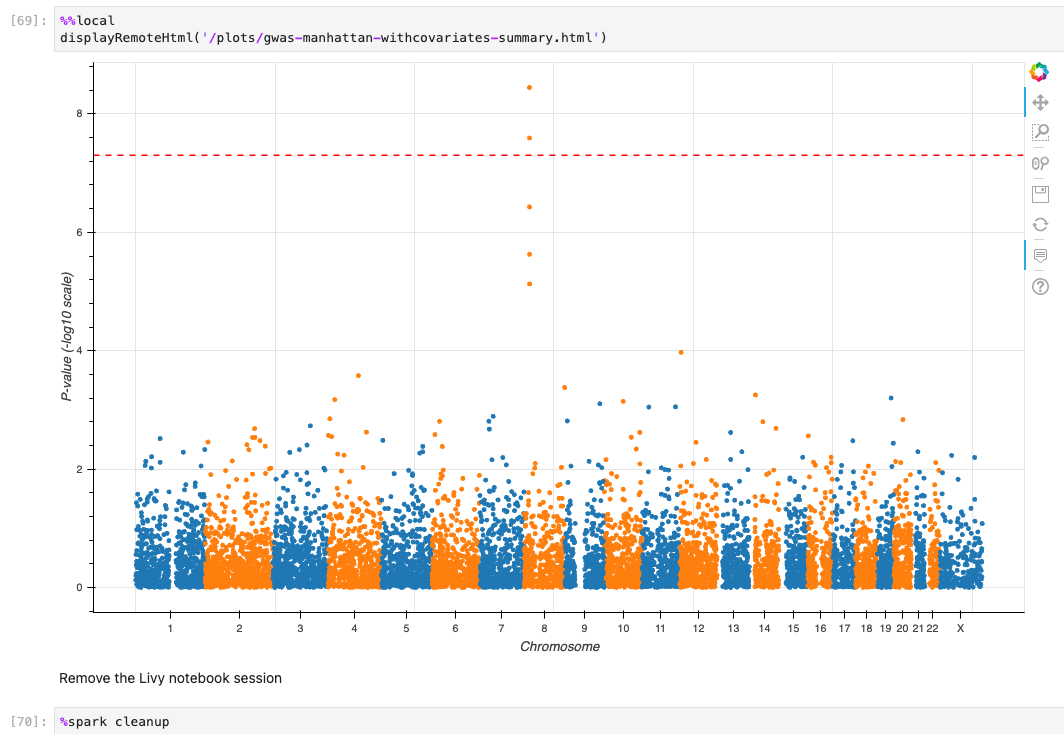
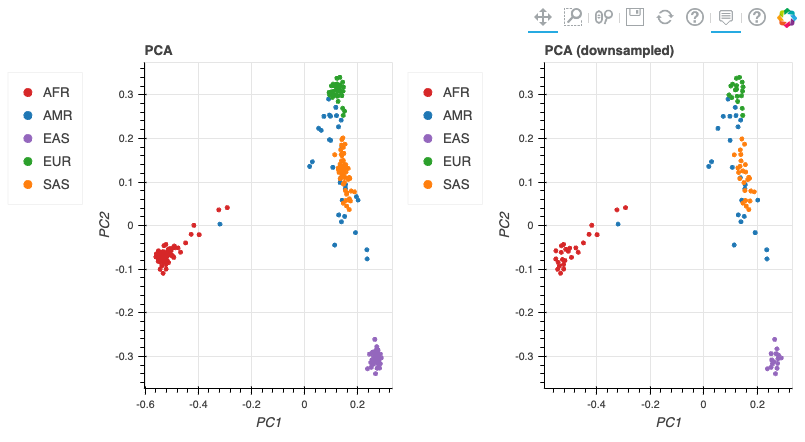
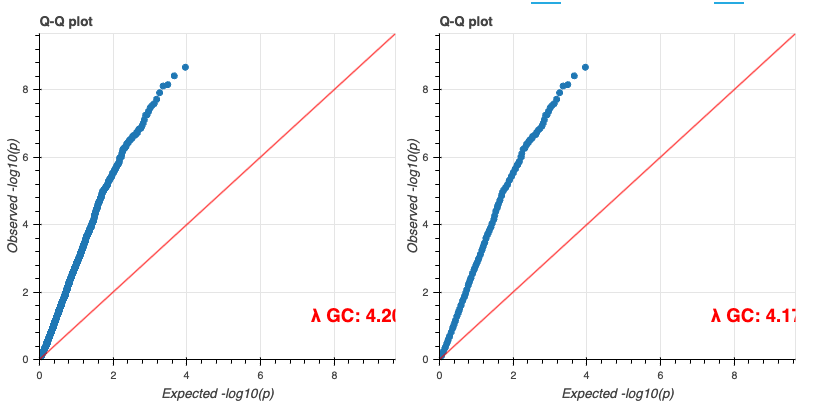
기타
VEP configuration
S3 버킷에서 해당 json파일 객체를 선택하고 Copy S3 URI를 클릭합니다.
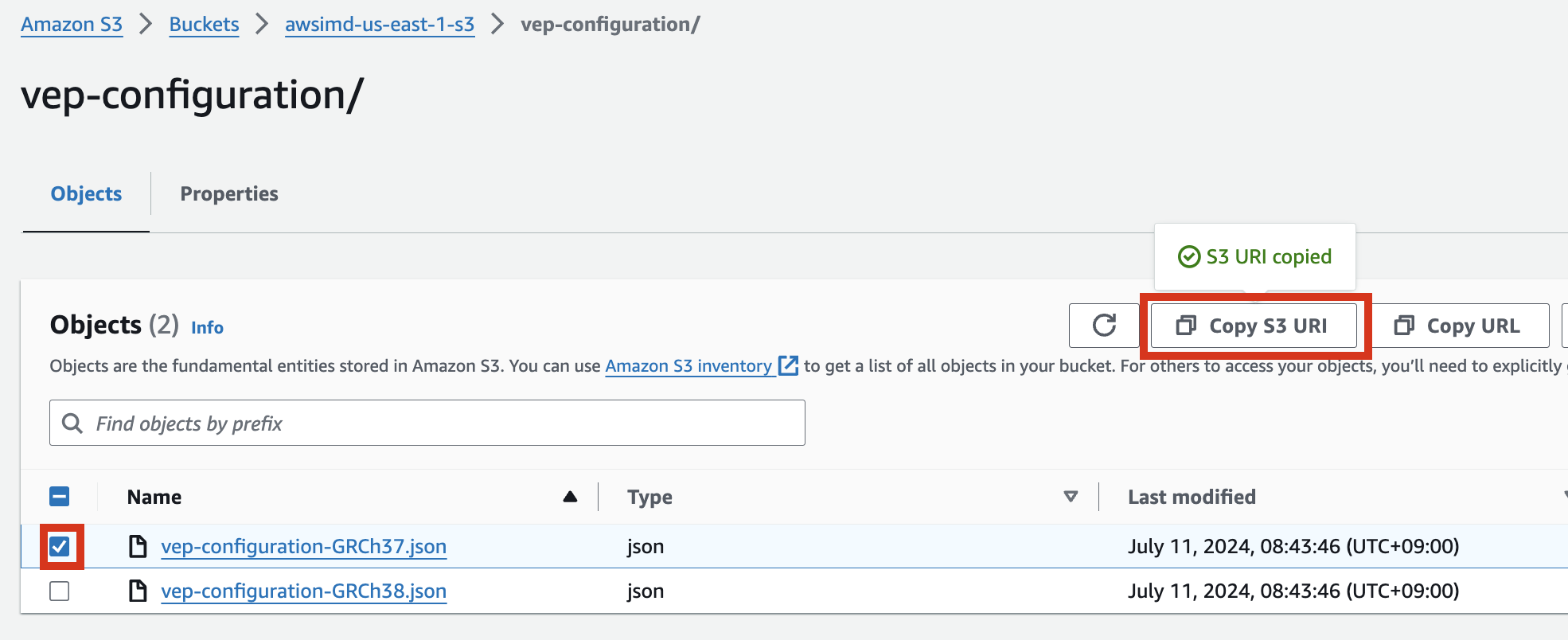
예시) vep-configuration-GRCh37.json
{
"command": [
"/opt/ensembl-vep/vep",
"--format", "vcf",
"--dir_plugins", "/opt/vep/plugins",
"--dir_cache", "/opt/vep/cache",
"--json",
"--everything",
"--allele_number",
"--no_stats",
"--cache", "--offline",
"--minimal",
"--assembly", "GRCh37",
"--plugin", "LoF,human_ancestor_fa:/opt/vep/loftee_data/human_ancestor.fa.gz,filter_position:0.05,min_intron_size:15,conservation_file:/opt/vep/loftee_data/phylocsf_gerp.sql,gerp_file:/opt/vep/loftee_data/GERP_scores.final.sorted.txt.gz",
"-o", "STDOUT"
],
"env": {
"PERL5LIB": "/opt/vep"
},
"vep_json_schema": "Struct{assembly_name:String,allele_string:String,ancestral:String,colocated_variants:Array[Struct{aa_allele:String,aa_maf:Float64,afr_allele:String,afr_maf:Float64,allele_string:String,amr_allele:String,amr_maf:Float64,clin_sig:Array[String],end:Int32,eas_allele:String,eas_maf:Float64,ea_allele:String,ea_maf:Float64,eur_allele:String,eur_maf:Float64,exac_adj_allele:String,exac_adj_maf:Float64,exac_allele:String,exac_afr_allele:String,exac_afr_maf:Float64,exac_amr_allele:String,exac_amr_maf:Float64,exac_eas_allele:String,exac_eas_maf:Float64,exac_fin_allele:String,exac_fin_maf:Float64,exac_maf:Float64,exac_nfe_allele:String,exac_nfe_maf:Float64,exac_oth_allele:String,exac_oth_maf:Float64,exac_sas_allele:String,exac_sas_maf:Float64,id:String,minor_allele:String,minor_allele_freq:Float64,phenotype_or_disease:Int32,pubmed:Array[Int32],sas_allele:String,sas_maf:Float64,somatic:Int32,start:Int32,strand:Int32}],context:String,end:Int32,id:String,input:String,intergenic_consequences:Array[Struct{allele_num:Int32,consequence_terms:Array[String],impact:String,minimised:Int32,variant_allele:String}],most_severe_consequence:String,motif_feature_consequences:Array[Struct{allele_num:Int32,consequence_terms:Array[String],high_inf_pos:String,impact:String,minimised:Int32,motif_feature_id:String,motif_name:String,motif_pos:Int32,motif_score_change:Float64,strand:Int32,variant_allele:String}],regulatory_feature_consequences:Array[Struct{allele_num:Int32,biotype:String,consequence_terms:Array[String],impact:String,minimised:Int32,regulatory_feature_id:String,variant_allele:String}],seq_region_name:String,start:Int32,strand:Int32,transcript_consequences:Array[Struct{allele_num:Int32,amino_acids:String,appris:String,biotype:String,canonical:Int32,ccds:String,cdna_start:Int32,cdna_end:Int32,cds_end:Int32,cds_start:Int32,codons:String,consequence_terms:Array[String],distance:Int32,domains:Array[Struct{db:String,name:String}],exon:String,gene_id:String,gene_pheno:Int32,gene_symbol:String,gene_symbol_source:String,hgnc_id:String,hgvsc:String,hgvsp:String,hgvs_offset:Int32,impact:String,intron:String,lof:String,lof_flags:String,lof_filter:String,lof_info:String,minimised:Int32,polyphen_prediction:String,polyphen_score:Float64,protein_end:Int32,protein_start:Int32,protein_id:String,sift_prediction:String,sift_score:Float64,strand:Int32,swissprot:String,transcript_id:String,trembl:String,tsl:Int32,uniparc:String,variant_allele:String}],variant_class:String}"
}
vep-tutorial 코드에서 아래 내용에서 위에서 복사한 S3 객체 URI로 수정하여 사용할 수 있습니다.
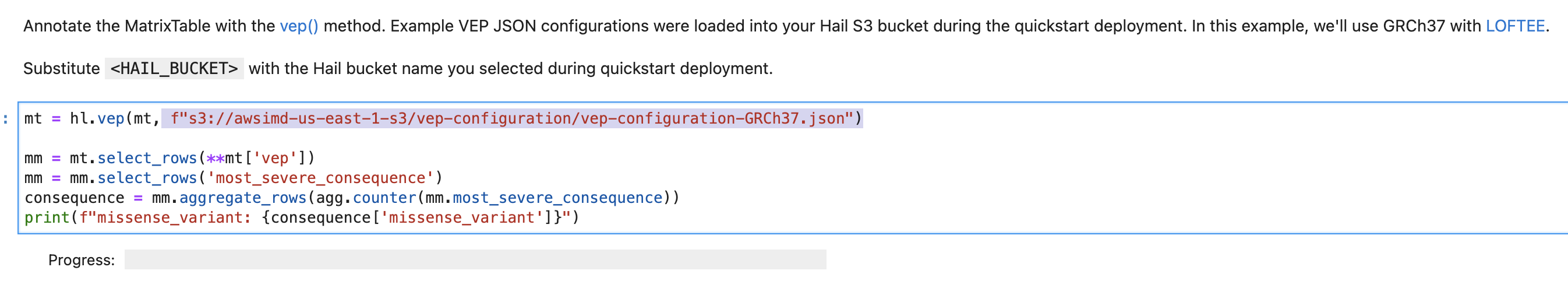
VEP 플러그인 설치
만일 VEP의 플러그인 설치에 변경사항 (추가 등)이 있다면 AMI를 다시 만들어야 합니다. 이 때 AMI를 만들 데 사용되는 VEP 설치에 관한 코드는 vep_install.sh 입니다. 해당 코드를 수정 후 다시 AMI를 빌드합니다.
다음을 참고하여 커스텀하게 Hail, VEP 툴을 설치 및 AMI를 빌드할 수 있습니다.
EMR 클러스터 EBS (HDFS) 동적 볼륨 늘리기
- 데이터가 클 경우 사전에 클러스터상에 구성된 볼륨의 용량이 부족할 수 있습니다. 아래 블로그 내용을 참고하여, EBS 볼륨의 부족분을 동적으로 늘릴 수 있습니다.
https://aws.amazon.com/ko/blogs/big-data/dynamically-scale-up-storage-on-amazon-emr-clusters/